INTRODUCTION
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is an essential therapeutic modality for many high-risk hematological diseases1. Successful outcomes after HSCT critically depend on the degree of donor-recipient matching at the human leukocyte antigen (HLA) loci; a poorly matched transplant will trigger a graft-versus-host disease (GVHD) and increase mortality2. The most frequent allogeneic transplants are those in which the donor is an HLA-identical sibling3,4; however, only 30% of patients who require a transplant have a compatible family donor, and with the reduction of children per family, this percentage is decreasing5. For patients without a suitable HLA-identical donor, a family-related donor as a source of hematoprogenitors for haploidentical-related hematopoietic stem cell transplantation (haplo-R-HSCT) represents a valid alternative1 with immediate donor availability in almost all patients6.
HLA-A, HLA-B, HLA-C, HLA-DRB1, and DQB1 (10/10) matching are optimum for hematopoietic stem cell transplant (HSCT)7; studies suggest that analysis of the HLA-DPB1 locus can be relevant for the success of haplo-HSCT due to its numerical importance, with over 1000 alleles described to date8. The HLA-DPB locus is located near the centromere of the chromosome 6 short arm9,10 and recombination at this point, which can give rise to a variation in offspring haplotype, frequently occurs10-12. In sibling donors matched at HLA-A, -B, and -DR, the rate of HLA-DPB1 mismatch has been estimated around 5% due to recombination13,14; complications as a result of this mismatching have been reported15. Thus, it is important to study the DPB1 locus in haplo-R-HSCT to confirm that the whole haplotype block is being transmitted as a unit12,16,17.
Complications in HSCT occur even in the setting of fully matched sibling transplantation18. A negative effect of incompatibility at the DPB1 locus in the unrelated HLA-identical HSCT setting has been reported15 and the previous studies have shown that the presence of DPB1 allele incompatibility resulted in significant differences in the incidence of a GVHD and disease relapse in unrelated HSCT, even if HLA 10/10 identity is present15. HLA-DP mismatches are relevant as it has been found that they participate in cellular and humoral HSC allograft rejection16.
The impact of HLA-DPB1 incompatibility in haplo-R-HSCT has not been established. The objective of the present study was to assess the rate and clinical effect of incompatibility at the HLA-DPB1 locus in the haplo-R-HSCT setting in a Hispanic cohort.
METHODS
Study population
We reviewed the clinical records and electronic files of 91 consecutive self-identified Hispanic patients with mostly malignant hematological diseases who underwent haplo-R-HSCT from January 2009 to October 2017 at the Hematology Department of the Dr. José E. González University Hospital, School of Medicine of the Universidad Autónoma de Nuevo León in Monterrey, Mexico. Transplants were performed in an outpatient setting after the administration of a reduced-intensity conditioning (RIC) regimen, as previously described3. Patients provided written informed consent. The Institutional Research and Ethics Committee approved the study protocol.
Haploidentical-related donor selection
Donors were healthy first-degree relatives selected according to standard criteria and availability. HLA compatibility was assessed by intermediate resolution molecular typing methods19. Donors were classified according to HLA matching with 5/10 antigens minimal compatibility. Allele designations were assigned according to the World Health Organization Nomenclature Committee for Factors of the HLA System20.
HLA-DPB1 allele determination
All subjects were typed at intermediate resolution for HLA-DPB1 by sequence-specific probe-based hybridization. Briefly, DNA was extracted from 300 µL of whole blood in ethylenediaminetetraacetic acid from donors and recipients using the automated Maxwell® 16 Blood DNA Purification System (Promega Corporation, Madison, WI). In the extracted DNA, polymerase chain reaction was carried out in a PROFLEX thermocycler (Applied Biosystems®, Foster City, CA) to amplify the region of the HLA-DP locus using the LABType SSO Class II DPA1 and DPB1® Typing Test (One Lambda, Canoga Park, CA). DNA amplification was carried out in a PROFLEX thermocycler (Applied Biosystems®, Foster City, CA) and the resulting product was transferred to a UNIPLATE® 96-well plate (Whatman GE, Healthcare Life Sciences, Madison, UK); the readings of each well and data acquisition were carried out in the LABScan™ 100 Luminex® 100 (Luminex Corporation Austin, TX). After reading, HLA Fusion™ software (One Lambda, Canoga Park, CA) was used to assign DPB alleles.
Mobilization and CD34+ hematoprogenitors harvest
Granulocyte colony-stimulating factor (G-CSF) at 10 µg/kg/day was administered subcutaneously for 5 days. CD34+ cells were collected with a Spectra Optia (Lakewood, CO) or a COBE Spectra (Gambro, Lakewood, CO) apheresis system and 5000-7000 mL of blood/m2 were processed in each apheresis to obtain ≥2 × 106 viable CD34+ cells/kg of the recipient’s body weight. CD34+ cells were measured by flow cytometry in a FACSCanto cytometer (Becton Dickinson, San Jose, CA).
Conditioning regimen for haplo-HSCT
A RIC scheme for adults included i.v. cyclophosphamide (Cy) 350 mg/m2/day and i.v. fludarabine (Flu) 25 mg/m2/day on days −5, −4, and −3 and i.v. melphalan 100 mg/m2/day on days −2 and −1. Infusion took place on day 0; i.v. Cy 50 mg/kg/day and Mesna 80% (Cy) on days +3 and +4; and oral cyclosporine A, 6 mg/kg/day and mycophenolate mofetil (MMF), 1 g/day on day +5 were administered for GVHD prophylaxis. In patients with a high risk of relapse, we used Cy 350 mg/m2/day and Flu 25 mg/m2/day on days −7, −6, and −5; busulfan (BU) 4 mg/kg/day on days −4, −3, and −2; and break on day −1. Infusion of the graft was performed on day 0; Cy 50 mg/kg/day and Mesna 80% (Cy) on days +3 and +4; cyclosporine A, 6 mg/kg/day and MMF 1 g/day on day +5; and G-CSF 300 mcg/day on days +7-+10. In patients with aplastic anemia, BU was not used, and each patient received conditioning according to their clinical status and transplant physician preference.
For children, conditioning consisted of a combination of Cy at 1500 mg/m2, Flu, 75 mg/m2, and i.v. BU, 9.6 mg/kg. As GVHD prophylaxis, patients received high-dose Cy (50 mg/kg) on days +3 and +4. Cyclosporine A, 6 mg/kg/d and MMF, 15 mg/kg 2 times per day were started on day +5. MMF was discontinued on day +35 and tapering of cyclosporine started on day +90.
Engraftment
Neutrophil engraftment was defined as an absolute neutrophil count ≥500/µL for 2 consecutive days and platelet engraftment as a count ≥20,000/µL for 2 consecutive days, at least 7 days from the last platelet transfusion21. Engraftment was also assessed by chimerism analysis by flow cytometry; in cases with a sex mismatch, a fluorescent in situ hybridization technique to demonstrate X or Y chromosome was used22. Complete donor host chimerism was defined by at least 95% donor cells and mixed chimerism by ≥5% recipient cells22,23. Studies were done on days 30 and 100 after haplo-R-HSCT. Primary graft failure was established by the absence of initial donor cell engraftment and if the patient never recovered from neutropenia, and secondary graft failure by loss of donor cells after initial engraftment24.
Statistical analysis
Data were analyzed using IBM SPSS Statistics for Windows v. 22.0 (IBM Corp., Armonk, NY). A descriptive analysis was performed; continuous variables were described as medians and ranges. The demographic and clinical characteristics were compared using the X2 test for categorical variables. The Kruskal–Wallis test was used for calculation of differences between variables and to compare data between groups. Overall survival (OS) was measured from the time of transplantation to time of death or last visit, with the Kaplan–Meier method and was compared using the logrank test. Cumulative incidence for relapse was measured from the time of transplantation to the time of relapse. Cumulative incidence of transplant-related mortality (TRM) was measured from the time of transplantation to the time of death without relapse/recurrence. TRM and relapse rates were estimated using cumulative incidence curves, taking competing events into consideration and compared with the Gray test. Binary logistic regression was used to evaluate the factors associated with an increased risk of presenting acute GVHD, and the Cox proportional hazard regression model with a 95% confidence interval (CI) (95% CI) was used to examine the association between the different variables and their effect on OS, TRM, and relapse. p < 0.05 was considered statistically significant.
RESULTS
Patients’ characteristics
Ninety-one patients with severe hematological diseases received a haploidentical (5/10 in HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1) related HSCT. Patient and donor demographics and transplantation characteristics, including donor-receptor family relationship, are shown in Table 1. Of the 91 donor-recipient pairs, 24 (26.37%) shared complete DPB1 identity, 60 (65.93%) had a mismatch at one allele, and 7 (7.70%) were mismatched at two alleles, indicating a high frequency of both homozygosity and recombination at the DPB1 locus. Only 6 (25%) of those homozygotes for DPB1 were also homozygotes for HLA-DR alleles and none shared complete identity for DQ. Among patients, the proportion of males was 60.4% and females, 39.6%. The median age for all patients was 18 years (range 0-64 years); mothers (38.5%) and fathers (20.9%) were the more frequent donors. For the fully matched HLA-DPB1 group, median age was 16 years (range 2-52 years) and 18.5 years for the 1-DPB1 mismatch group (range 0-64 years). The 2-DPB1 mismatch group had a median age of 19 years (range 3-48 years); there was no statistical difference in age.
Table 1 Characteristics of 91 patients who underwent haploidentical-related hematopoietic stem cell transplant from 2009 to 2017 at "Dr. José E. González" University Hospital in Monterrey, Mexico, according to HLA-DPB1 matching
| Characteristic | Haploidentical (n=91) | 0-DPB1 mismatch (n=24) | 1-DPB1 mismatch (n=60) | 2-DPB1 mismatch (n=7) | p |
|---|---|---|---|---|---|
| Age, median (range) | 18 (0-64) | 16 (2-52) | 18.5 (0-64) | 19 (3-48) | 0.351 |
| <16 years (%) | 36 (39.6) | 12 (50) | 21 (35) | 3 (42.9) | 0.439 |
| >16 years (%) | 55 (60.4) | 12(50) | 39 (65) | 4 (57.1) | 0.439 |
| Patient gender (%) | |||||
| Male | 55 (60.4) | 14 (58.3) | 35 (58.3) | 6 (85.7) | 0.363 |
| Female | 36 (39.6) | 10 (41.7) | 25 (41.7) | 1 (14.3) | |
| Diagnosis (%) | |||||
| ALL | 42 (46.2) | 13 (54.2) | 24 (40) | 5 (71.4) | 0.610 |
| AML | 16 (17.6) | 4 (16.7) | 11 (18.3) | 1 (14.3) | |
| AA | 6 (6.6) | 2 (8.3) | 4 (6.7) | 0 (0) | |
| NHL | 6 (6.6) | 1 (4.2) | 5 (8.3) | 0 (0) | |
| HL | 6 (6.6) | 0 (0) | 6 (10) | 0 (0) | |
| MDS | 3 (3.3) | 2 (8.3) | 1 (1.7) | 0 (0) | |
| CML | 6 (6.6) | 1 (4.2) | 4 (6.7) | 1 (14.3) | |
| CLL | 2 (2.2) | 0 (0) | 2 (3.3) | 0 (0) | |
| SCID | 3 (3.3) | 0 (0) | 3 (5) | 0 (0) | |
| Other | 1 (1.1) | 1 (4.2) | 0 (0) | 0 (0) | |
| Donor/recipient sex match (%) | |||||
| Male to male | 25 (27.5) | 6 (25) | 15 (25) | 4 (57.1) | 0.187 |
| Male to female | 13 (14.3) | 4 (16.7) | 8 (13.3) | 1 (14.3) | 0.925 |
| Female to male | 31 (34.1) | 8 (33.3) | 21 (35) | 2 (28.6) | 0.940 |
| Female to female | 22 (24.2) | 6 (25) | 16 (26.7) | 0 (0) | 0.295 |
| Family relationship (%) | |||||
| Brother | 16 (17.6) | 4 (16.7) | 10 (16.7) | 2 (28.6) | 0.729 |
| Sister | 16 (17.6) | 4 (16.7) | 11 (18.3) | 1 (14.3) | 0.956 |
| Mother | 35 (38.5) | 9 (37.5) | 25 (41.7) | 1 (14.3) | 0.368 |
| Father | 19 (20.9) | 5 (20.8) | 11 (18.3) | 3 (42.9) | 0.319 |
| Son | 3 (3.3) | 1 (4.2) | 2 (3.3) | 0 (0.0) | 0.862 |
| Daughter | 2 (2.2) | 1 (4.2) | 1 (1.7) | 0 (0.0) | 0.716 |
| CMV status (R/D) (%) | |||||
| Positive/positive | 51 (56) | 14 (58.3) | 33 (55) | 4 (57.1) | 0.916 |
| Positive/negative | 9 (9.9) | 2 (8.3) | 5 (8.3) | 2 (28.6) | 0.227 |
| Negative/negative | 6 (6.6) | 1 (4.2) | 5 (8.3) | 0 (0) | 0.601 |
| Negative/positive | 6 (6.6) | 3 (12.5) | 3 (5) | 0 (0) | 0.350 |
| Unknown | 19 (20.9) | 4 (16.7) | 14 (23.3) | 1 (14.3) | 0.719 |
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; AA, aplastic anemia; MDS, myelodysplastic syndrome; NHL, non-Hodgkin lymphoma; HL, Hodgkin lymphoma; CML, chronic myeloid leukemia; CLL, chronic lymphocytic leukemia; CSID, combined severe immunodeficiency; CMV, cytomegalovirus; R/D, recipient/donor.
The most frequent diagnoses were acute lymphoblastic leukemia (ALL) in 42 (46.2%) patients, acute myeloid leukemia (AML) 16 (17.6%), non-Hodgkin lymphoma (NHL) 6 (6.6%), Hodgkin’s lymphoma (HL) 6 (6.6%), and aplastic anemia (AA) in 6 (6.6%). The most frequent donor/recipient sex match was female to male in 31 (34.1%) followed by male to male in 25 (27.5%) donor-recipient pairs. More frequent CMV status was positive-positive in 51 (56%) pairs and was not statistically significant (Table 1). Regarding ABO blood type donor-recipient compatibility, 71 grafts (78%) were matched pairs, 11 (12.1%) minor mismatched, and 9 (9.9%) major mismatched transplants.
There were 36 (39.6%) deaths: 15 (41.7%) secondary to baseline disease, including 10 with ALL, 2 AML, 1 NHL, 1 CML, and 1 aplastic anemia; 14 (38.9%) were related to sepsis; 3 (8.3%) to gastrointestinal bleeding; and 4 (11.1%) to acute kidney failure with a severe hydroelectrolytic imbalance (n = 2) and multiple organ dysfunction syndrome (n=2).
HLA-DPB1 alleles
Twenty-four different HLA-DPB1 alleles were found in patients who received a haplo-R-HSCT; the most frequent were DPB1*04:01 (34.1%) and DPB1*04:02 (27.5%). Other alleles are shown in table 2.
Table 2 Frequency of HLA-DPB1 alleles in patients who received a haploidentical transplant and their family donors
| Allele | Number | Percentage |
|---|---|---|
| 04:01 | 62 | 34.1 |
| 04:02 | 50 | 27.5 |
| 03:01 | 14 | 7.7 |
| 02:01 | 10 | 5.5 |
| 05:01 | 6 | 3.3 |
| 70:01:00 | 5 | 2.7 |
| 01:01 | 3 | 1.6 |
| 13:01 | 3 | 1.6 |
| 17:01 | 3 | 1.6 |
| 65:01:00 | 3 | 1.6 |
| 105:01:00 | 3 | 1.6 |
| 424:01:00 | 3 | 1.6 |
| 14:01 | 2 | 1.1 |
| 22:01 | 2 | 1.1 |
| 57:01:00 | 2 | 1.1 |
| 126:01:00 | 2 | 1.1 |
| 621:01:00 | 2 | 1.1 |
| 02:02 | 1 | 0.5 |
| 10:01 | 1 | 0.5 |
| 11:01 | 1 | 0.5 |
| 94:01:00 | 1 | 0.5 |
| 131:01:00 | 1 | 0.5 |
| 155:01:00 | 1 | 0.5 |
| 410:01:00 | 1 | 0.5 |
| Total | 182 | 100 |
Clinical outcomes and main complications
The median of days for myeloid and platelet engraftment was 16 and 17 for the whole cohort, with no statistical difference between the groups. Chimerism analysis was carried out, and its main results are shown in table 3. Sixty-seven (73.6%) patients presented fever and neutropenia in the post-transplant period, whereas infections developed in 59 (64.8%) recipients; both tended to be higher in the 1-DPB1 mismatched group. For the 2-mismatch group, no statistical differences in clinical outcomes were found.
Table 3 Clinical outcomes according to HLA-DPB1 matching of 91 Hispanic patients with diverse hematologic diseases who received a haploidentical-related transplant from 2009 to 2017 in Northeast Mexico
| All patients (n=91) | 0 mismatched DPB1 (n=24) | 1 mismatched DPB1 (n=60) | 2 mismatched DPB1 (n=7) | p | |
|---|---|---|---|---|---|
| Engraftment days (median, range) | |||||
| Myeloid | 16 (10-56) | 16 (10-56) | 16 (11-43) | 16.5 (14-24) | 0.678 |
| Platelets | 17 (9-56) | 17 (10-56) | 17 (9-30) | 18.5 (12-24) | 0.542 |
| Chimerism (%) 30 days | |||||
| Complete chimerism | 53 (58.2) | 13 (54.2) | 37 (61.7) | 3 (42.8) | 0.429 |
| Mixed chimerism | 12 (13.2) | 5 (20.8) | 5 (8.3) | 2 (28.6) | 0.745 |
| Not available | 263 (28.6) | 6 (25) | 18 (30) | 2 (28.6) | 0.216 |
| 100 days | |||||
| Complete chimerism | 37 (40.7) | 8 (33.3) | 25 (41.7) | 4 (57.1) | 0.378 |
| Mixed chimerism | 8 (8.8) | 4 (16.7) | 3 (5) | 1 (14.3) | 0.076 |
| Not available | 46 (50.5) | 12 (50) | 32 (53.3) | 2 (28.6) | 0.533 |
| Complications after HSCT (%) | |||||
| Fever and neutropenia | 67 (73.6) | 15 (62.5) | 47 (78.3) | 5 (71.4) | 0.328 |
| Infections (any type) | 59 (64.8) | 15 (62.5) | 40 (66.7) | 4 (57.1) | 0.849 |
| Transfusions | 64 (70.3) | 16 (66.7) | 42 (70) | 6 (85.7) | 0.621 |
| Mucositis (I-IV) | 20 (22.0) | 6 (25) | 12 (20) | 2 (28.6) | 0.802 |
| CMV PCR (+) | 14 (15.4) | 5 (20.8) | 7 (11.7) | 2 (28.6) | 0.347 |
| Acute GVHD | 32 (35.2) | 7 (29.2) | 21 (35) | 4 (57.1) | 0.394 |
| Grade I | 13 (14.3) | 2 (8.3) | 9 (15) | 2 (28.6) | 0.758 |
| Grade II | 14 (15.4) | 4 (16.7) | 8 (13.3) | 2 (28.6) | 0.593 |
| Grade III | 6 (6.6) | 1 (4.2) | 5 (8.3) | 0 (0) | 0.531 |
| Grade IV | 1 (1.1) | 0 (0) | 1 (1.7) | 0 (0) | 0.773 |
| Chronic GVHD | 21 (23.1) | 5 (20.8) | 14 (23.3) | 2 (28.6) | 0.910 |
| Relapse | 33 (36.3) | 9 (37.5) | 20 (33.3) | 4 (57.1) | 0.459 |
| Engraftment failure | 18 (19.8) | 7 (29.2) | 9 (15) | 2 (28.6) | 0.281 |
| Death | 36 (39.6) | 8 (33.3) | 26 (43.3) | 2 (28.6) | 0.577 |
Transfusion of blood products was required in 64 (70.3%) patients and tended to be higher in the HLA-DPB1 unmatched group. Engraftment failure was documented in 18 patients (19.8%), tending to be higher in the fully matched HLA-DPB1 allele group, with no statistical significance reached. Other salient clinical characteristics are displayed in table 3. Acute GVHD developed in 32 (35.2%) patients; 7 in the 0-DPB1 mismatch group, 21 in the 1-DPB1 mismatch, and 4 in the 2-DPB1 mismatch group; Grade 1 and 2 of acute GVHD developed more frequently in the unmatched group (Table 3). Binary logistic regression analysis did not show risk factors associated with this complication (Table S1).
Table S1 Binary logistic regression for factors associated with acute GVHD and Cox regression analysis for overall survival in 91 allografted patients with hematologic disease who received a haploidentical-related stem cell transplant in Monterrey, Mexico
| Acute GVHD | Overall survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||
| R (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Sex (male) | ||||||||
| Female | 1 | 1 | 1 | 1 | ||||
| Male | 0.92 (0.37-2.32) | 0.876 | 1.09 (0.40-2.97) | 0.862 | 1.03 (0.52-2.03) | 0.933 | 0.85 (0.42-1.75) | 0.678 |
| Age | 1.00 (0.97-1.03) | 0.934 | 0.99 (0.96-1.03) | 0.828 | 1.00 (0.98-1.02) | 0.841 | 0.98 (0.95-1.01) | 0.295 |
| Mismatched at HLA DPB1 | ||||||||
| 0 mismatched allele | 1 | 1 | 1 | 1 | ||||
| 1 mismatched allele | 1.30 (0.46-3.65) | 0.609 | 1.10 (0.36-3.31) | 0.857 | 1.38 (0.62-3.05) | 0.426 | 1.08 (0.44-2.63) | 0.853 |
| Dose CD34+ cells/Kg | 1.04 (0.92-1.17) | 0.467 | 1.04 (0.92-1.19) | 0.482 | 0.82 (0.73-0.92) | 0.001 | 0.81 (0.71-0.91) | 0.001 |
| Infections | ||||||||
| Yes | 1.50 (0.56-4.00) | 0.419 | 1.39 (0.48-4.02) | 0.543 | 0.87 (0.42-1.79) | 0.705 | 0.62 (0.29-1.34) | 0.232 |
| No | 1 | 1 | 1 | 1 | ||||
| Transfusions | ||||||||
| Yes | 0.56 (0.21-1.47) | 0.245 | 0.48 (0.17-1.36) | 0.173 | 1.71 (0.77-3.79) | 0.184 | 1.44 (0.63-3.32) | 0.382 |
| No | 1 | 1 | 1 | 1 | ||||
| Fever and neutropenia | ||||||||
| Yes | 2.00 (0.65-6.16) | 0.224 | 1.74 (0.50-6.02) | 0.377 | 1.09 (0.49-2.42) | 0.823 | 0.95 (0.41-2.18) | 0.911 |
| No | 1 | 1 | 1 | 1 | ||||
Mortality
Thirty-six (39.6%) patients died; 8 (8.80%) belonged to the fully matched HLA-DPB1 group, and 26 (28.6%) to the 1-mismatch group, with no statistical difference (p = 0.577). The remaining 2 (2.20%) patients who died belonged to the 2-mismatched alleles DPB1 group. The median OS was 19 months for the 1-DPB1 mismatch group; for the fully matched and unmatched groups, median OS was not reached. Two-year OS was 51.3 ± 6.8% for all haplo-related transplanted patients; 59.9 ± 11.6% for the 0-DPB1 mismatch group, 46.4% (44.70-48.07%) for the 1-DPB1 mismatch group, and 55.6 ± 24.8% for the unmatched HLA-DPB1 group (p = 0.409) (Figs. 1a and b). In Cox regression analysis, only the dose of CD34+ cells was a protective factor for OS (Table S1).
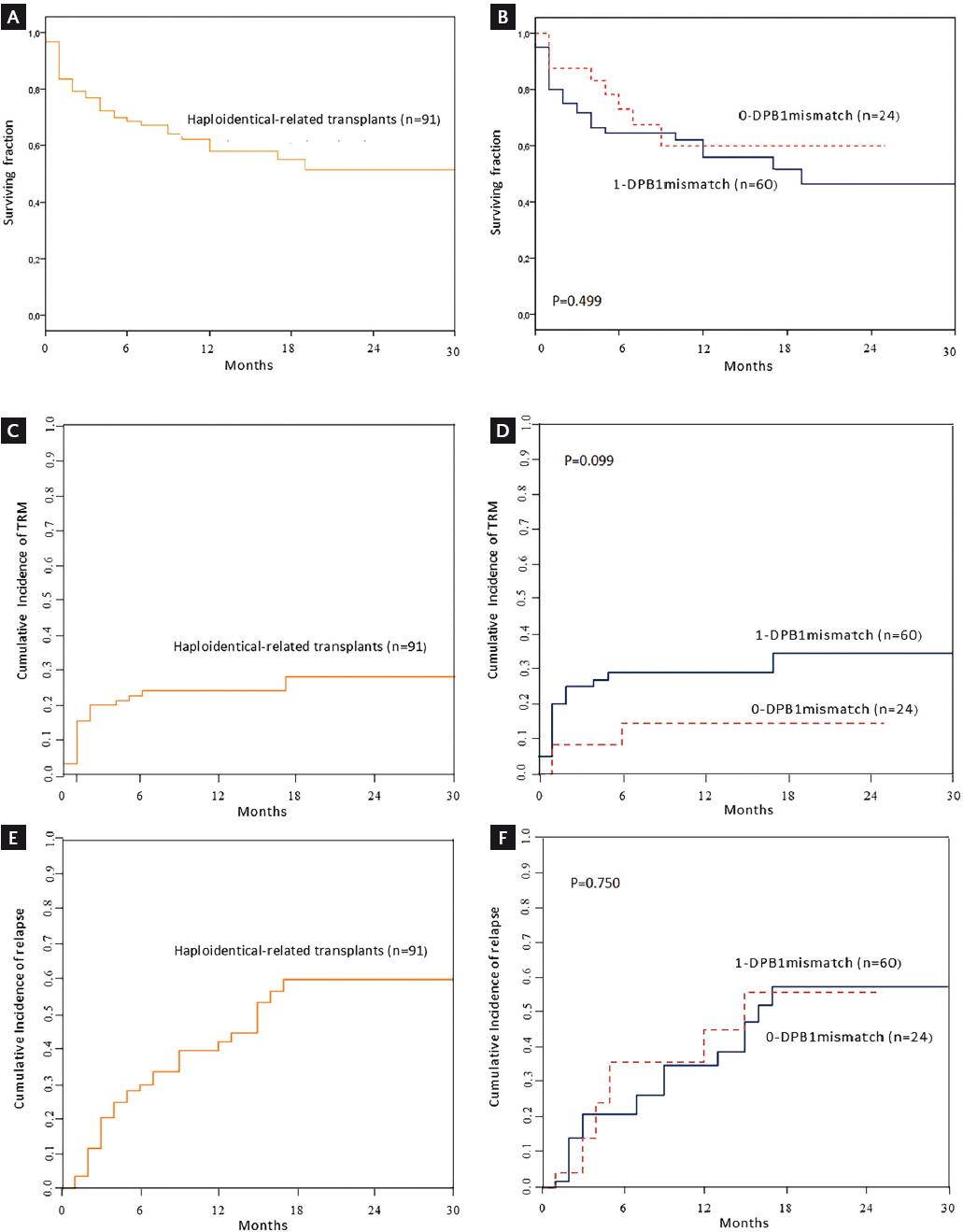
Figure 1 (A) OS for 91 haploidentical-related hematopoietic stem cell recipients receiving reduced-intensity conditioning. (B) Comparison of OS between patients fully matched and partially matched at the HLA-DPB1 allele. (C) Cumulative incidence of TRM for haploidentical-related hematopoietic stem cell transplants in 91 patients. (D) Comparison of cumulative incidence for TRM between patients fully matched and partially matched at the HLA-DPB1 allele. (E) Cumulative incidence of relapse in 91 haploidentical-related hematopoietic stem cell allografted patients following reduced-intensity conditioning. (F) Comparison of cumulative incidence for relapse between patients fully matched and partially matched at the HLA-DPB1 allele. OS, overall survival; TRM, transplant-related mortality.
The cumulative incidence of TRM at 2 years for all patients was 28 ± 6%; for the 0-DPB1 mismatch group, it was 14 ± 8.1% versus 34 ± 7.8% for the 1-DPB1 mismatch group (p = 0.099). The data, shown in Figs. 1c and d, suggest that a significant difference might be reached by increasing the sample size. In the unmatched group, one of two deaths was due to TRM.
Relapse
Cumulative incidence of relapse at 2 years was 60 ± 7.8% for all haplo-R-HSCT, 56 ± 15.3% for the 0-DPB1 mismatch, and 58 ± 10% in the 1-DPB1 mismatch group, with no significant difference (p = 0.750) (Figs. 1e and f). For the unmatched group, relapse developed in 4 of 7 (57.1%) patients. No risk factors were identified in Cox regression analysis of the cumulative incidence of relapse and TRM (Table S2).
Table S2 Cox regression analysis of cumulative incidence of relapse and transplant-related mortality (TRM) for 91 haploidentical-related transplants in hematologic patients of a Hispanic group in Northeast Mexico
| TRM | Relapse | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Sex (male) | ||||||||
| Female | 1 | 1 | 1 | 1 | ||||
| Male | 1.95 (0.77-4.92) | 0.160 | 1.81 (0.68-4.80) | 0.230 | 0.52 (0.25-1.08) | 0.080 | 0.47 (0.20-1.14) | 0.095 |
| Age | 1.01 (0.98-1.04) | 0.460 | 1.05 (0.97-1.03) | 0.720 | 0.99 (0.97-1.02) | 0.550 | 1.00 (0.97-1.03) | 0.770 |
| Mismatched at HLA DPB1 | ||||||||
| 0 mismatched allele | 1 | 1 | 1 | 1 | ||||
| 1 mismatched allele | 2.57 (0.79-8.32) | 0.120 | 2.90 (0.86-9.82) | 0.086 | 0.88 (0.41-1.87) | 0.740 | 0.85 (0.38-1.90) | 0.700 |
| CD34+<5 x 106/Kg | 0.99 (0.97-1.01) | 0.450 | 0.99 (0.97-1.01) | 0.410 | 0.99 (0.97-1.01) | 0.480 | 0.99 (0.97-1.02) | 0.670 |
| Infections | ||||||||
| Yes | 0.60 (0.26-1.38) | 0.240 | 0.62 (0.27-1.43) | 0.270 | 0.96 (0.44-2.11) | 0.930 | 1.00 (0.41-2.40) | 0.990 |
| No | 1 | 1 | 1 | 1 | ||||
| Transfusions | ||||||||
| Yes | 1.59 (0.63-4.02) | 0.330 | 1.32 (0.52-3.36) | 0.560 | 1.30 (0.60-2.78) | 0.500 | 1.34 (0.58-3.11) | 0.490 |
| No | 1 | 1 | 1 | 1 | ||||
| Fever and neutropenia | ||||||||
| Yes | 1.10 (0.42-2.88) | 0.850 | 0.88 (0.32-2.44) | 0.820 | 0.80 (0.36-1.78) | 0.590 | 0.71 (0.27-1.84) | 0.490 |
| No | 1 | 1 | 1 | 1 | ||||
DISCUSSION
The major limitations for haplo-HSCT allografting are the high incidence of TRM, graft failure, GVHD, and poor OS rates25. Nevertheless, for patients who lack an HLA-identical sibling, the haplo-R-HSC transplant modality represents a potentially life-saving alternative. HLA-matched sibling donors share both alleles of HLA-A, -B, -C, -DR, -DQ, and -DP (12/12). Exceptions do occur in 1-5% of cases, accounted for by genomic recombination, with the highest frequency reported for HLA-DP due to at least one recombination hotspot between DP and DQ26-28. HLA polymorphisms represent a barrier to HSC transplantation because HLA incompatibilities at the allele level can be recognized by alloreactive T lymphocytes29. HLA-DPB1 is the first locus explored as a model for clinically permissive donor-recipient HLA mismatches, and this has led to increased interest into HLA-DP role in unrelated HSCT. Thus, some HLA-DPB1 mismatches are considered permissive when the expressed T-cell epitope structure is similar in donor-recipient pairs, while others are considered non-permissive with greater differences in T-cell epitope structure, which may put the recipient at increased risk for suboptimal outcomes30. A negative effect of DPB1 locus incompatibility in unrelated HLA-identical HSCT has been documented31. Nevertheless, the relevance of HLA-DPB1 compatibility in the haplo-R-HSCT setting has not been established. Thus, we assessed the impact of incompatibility at this locus on clinical outcomes after related haplo-HSCT in a Hispanic cohort.
Importantly, HLA-DPB1 typing results in our cohort showed that 24 of 91 patients did not have allelic differences in HLA-DPB1, and therefore, 60, two-thirds, were true HLA-DPB1 haplo-R-HSCT. This reflects a high degree of homozygosity among the self-identified Hispanic individuals of the study population, and frequent recombination at this locus, since seven patients encoded no common DPB1 allele. Interestingly, only a quarter of HLA-DPB1 homozygous were also homozygous for HLA-DR and none shared both DQ alleles.
The most frequent HLA-DPB1 alleles found were DPB1*04:01 (34.1%) and DPB1*04:02 (27.5%). Studies in the United States report a frequency for DPB1*04:01 allele from 10.4% to 38.90% and 12% to 62.0% for DPB1*04:0232-35. In a cohort of unrelated European American stem cell donors, this frequency was 43.8% for DPB1*04:01 and 11.5% DPB1*04:0236. For Latin-American countries, DPB1*04:02 allele frequencies range from 13.2% to 89.10%, whereas for the DPB1*04:01 allele, it is 0% to 15.3%34,37-42, reflecting considerable expression heterogeneity of HLA molecules among populations across the continent.
Infections are a common problem after haplo-HSCT and account for substantial morbidity and mortality42. Fever and neutropenia and the need for transfusion in the post-transplant period were the main complications in our haplo-R-HSCT cohort, similar to the studies that found a higher incidence of these complications in the haploidentical than in HLA-identical transplants43.
Acute GVHD affects 10-50% of HSCT recipients even with the use of standard prophylactic immunosuppressive regimens44. It developed in 35.2% of our recipients and tended to be higher in mismatched than in HLA-DPB1 fully matched recipients, although the difference was not statistically significant. Grade 1 and 2 acute GVHD tended to develop in the mismatched group, with no risk factor associated with an increased incidence of this complication. No significant difference according to DPB1-allele matching was found in chronic GVHD; nevertheless, this complication tended to be higher in the one-mismatch group.
Relapse is the main cause of treatment failure after allogeneic HSCT45 and the principal cause of death 100 days after HLA-identical sibling and unrelated allografting46. One study found that the hazard for relapse between patients matched at -A,-B,-C, -DR, and -DQ alleles and mismatched at HLA-DPB1 was lower than for patients matched at DPB1, reflecting the importance of the graft versus leukemia effect47. In contrast, in patients mismatched for at least one other HLA allele, the impact of DPB1 mismatch was not significant48. In our haplo-R-HSCT cohort, cumulative incidence of relapse at 2 years was 60%, similar to the general rate previously reported3. In another study in patients with newly diagnosed AML that received a haplo-unrelated transplant, the cumulative incidence of relapse at 3 years was 70%49. In our group, patients matched at the HLA-DPB1 allele had a relapse rate of 56% versus 58% in the mismatched group at 2 years; in contrast, in studies on unrelated haploidentical transplants mismatched at HLA-DPB1, a significant difference in relapse rate between these groups was found15,50.
Haplo-HSCT leads to a higher incidence of TRM and overall mortality compared with HLA-identical sibling HSCT43. OS in our study was 51.3%, similar to 58% in another report51. Importantly, although no statistical difference between fully and partially compatible recipients at the HLA-DPB1 allele was documented, fully matched recipients had a considerably lower TRM. This strongly suggests that a significant difference might be reached by increasing the number of patients studied; therefore, statistically powered cooperative studies are needed to answer this question. Remarkably, the only risk factor statistically associated with an increased risk of mortality was a lower dose of CD34+ cells.
In our cohort, TRM at 1 year was 24%; a recent systematic review and meta-analysis found 5%-42% rate after 12 months52. For fully and partially HLA-DPB1 matched patients, no difference in TRM or any risk factor associated with higher mortality existed, which supports recent observations that HLA-DP antigens can be a model for clinically permissive mismatches eliciting limited T-cell alloreactivity16. Importantly, this is the first study to analyze the clinical relevance of compatibility at the HLA-DPB1 locus in patients who receive a haplo-R-HSCT in an ambulatory setting after RIC.
It is relevant to underscore than in pediatric populations, and more so in those from low- and middle-income countries, finding a complete match for major histocompatibility complex haplotypes is considerably more difficult. This has stimulated the development of alternative donor sources, including haploidentical grafts, despite this type of haplo-HSCT carries a higher risk for graft failure and GVHD. In this respect, post-transplant Cy administration has resulted in improved outcomes, helping to advance the field of pediatric haplotransplantation53,54.
Some limitations of our study should be noted, including its retrospective design and the limited number of allografted patients; prospective, larger studies in different populations are needed to confirm these findings.
In conclusion, mismatching for DPB1 did not lead to significant differences for main transplant outcomes in a cohort suffering severe hematologic disease, and mismatching at this locus was clinically tolerable.
SUPPLEMENTARY DATA
Supplementary data are available at Revista de Investigación Clínica online (www.clinicalandtranslationalinvestigation.com). These data are provided by the corresponding author and published online for the benefit of the reader. The contents of supplementary data are the sole responsibility of the authors.











 nueva página del texto (beta)
nueva página del texto (beta)


