1 Introduction
Amazon biome is one of the main sources of biodiversity in the world's ecosystems [1]. Wildlife is an important component within its territory [2, 3]. While the species list is growing all the time, only a fraction of the Amazon’s enormous biodiversity is known to science [4, 5].
According to estimates only 90-95 per cent of mammals, birds and plants are known, only 2-10 per cent of insects have been described, and only 2,500 from the approximately 6,000 – 8,000 amazon fish species have been described [6]. Eight countries share responsibility for the Amazon, one of them is Peru which is home of 11.27 per cent of the biome. Peru stands out as one of the most biodiverse countries in the world.
The extensive Amazonian forests cover 62% of the Peruvian territory and they are home to approximately 50% of the plant species registered by the Ministry of Environment-MINAM [7]. This remarkable diversity also includes numerous wildlife endemic species in the region.
For example, 115 endemic bird species have been identified (representing 6% of the world total), 109 mammal species (27.5% of the world total), 185 amphibian species (48.5% of the world total) and 59 endemic butterfly species (12.5% of the world population) [8].
The efforts to conservate and manage fauna in the Amazon Biome does not still fill gaps of knowledge about tropical fauna [3, 9]. It is needed to strength accurate identification and monitoring of wildlife through discovering and documentation. Tradition-ally the identification is based on different biological assessments of biodiversity.
However, identification and documentation methods of wildlife in the Amazon represents a considerable challenge for professionals engaged in population biology and ecology studies, as it involves a high cognitive load and significant time consumption [10]. This difficulty is attributed to the existence of multiple types of animals that exhibit high similarity to each other, making difficult their precise classification [11].
Over the past few decades, automated species identification has brought about a revolution in conventional methodologies [12]. Recent research has demonstrated the emergent use of artificial intelligence (AI) and more specifically computer vision in the identification and monitoring biodiversity species, is the case of MobileNetV3 a deep learning model that were successfully used enabling faster and more efficient analysis at identifying mangrove species [13].
The same technology has been used to detect plant species [14]. Similar technologies have contributed to detect camels on roads [15], as well as to identify rodent species [16]. Another example is the use of AlexNet model to identify ringed seal of the Saimaa, according to results the experiment get an accuracy of 91.2% in the individual identification of species [17].
In the same line, [18] proposed a framework for animal’s recognition which consisting of 2 Convolutional neural network (CNN) based models for image classification, results show values close to 90% in the identification of 3 most common animals. [19] proposed the application of a 3-branch VGG CNN in parallel with the aim of recognizing wild animal species. CNN has contributed to recognize wild boars [20] toads/frogs, lizards and snakes [21].
The transfer learning architecture called YOLOv3 has enabled the wildlife monitoring [11]. The advantages offered by CNN’s applications in image recognition have been applied for different purposes [22, 23, 24], constructed the wildlife dataset of Northeast China Tiger and Leopard National Park to identify and process images captured with camera traps. Using three deep learning object detection models YOLOv5, FCOS and Cascade R-CNN, they found an average Accuracy of 97.9%, along with an approximate mAP50 of 81.2% for all three models. Along the same lines, [25] used a transfer learning approach to detect the presence of four endangered mammals in the forests of Negros Island (Viverra tangalunga, Prionailurus javanensis sumatranus, Rusa alfredi and Sus cebifron).
The authors used the YOLOv5 model as a detection method. The trained model yielded a mean mAP50 of 91%. [26] proposed the identification of snake, lizard and toad/frog species from camera trap images using CNN. The results obtained by accuracy were 60% in the validation stage. [27] proposed a system to detect animals on the road and avoid accidents.
To do this, the animals were classified into groups of capybaras and donkeys. The authors used two variants of pre-trained CNN models: Yolov4 and Yolov4-tiny.
The results showed an accuracy of 84.87% and 79.87% for Yolov4 and Yolov4-tiny, respectively. As shown, there are important advances at animal detection using deep learning technologies.
However, they have not been applied to detect wildlife species in the Peruvian part of the Amazon biome yet. The main objective of this study was to evaluate the application of the YOLO algorithm in its versions YOLOv5x6, YOLOv5l6, YOLOv7-W6, YOLOv7-E6, YOLOv8I and YOLOv8x in the detection of wildlife species in the Peruvian Amazon.
To achieve this, we have evaluated the aforementioned models using the following metrics: Precision, Recall, F1-Score and mAP50, applied to six species: Ara ararauna, Ara chloropterus, Ara macao, Opisthocomus hoazin, Pteronura brasiliensis and Saimiri sciureus.
In addition, as part of our contribution to the scientific community, we provide a labelled dataset for classification and/or detection of these species. We have structured the remaining contents of the paper as follows: In Section 2, the methodology adopted to conduct the experiments is presented in detail.
Results and discussions are addressed in Section 3, while our conclusions are presented in Section 4.
2 Material and Methods
We performed our experiments using the machine learning technique called transfer learning, which consists of using previously learned knowledge trained on large volumes of public images [28, 29]. Specifically, we have used the object detection algorithm in images and video called YOLO (You Only Look Once) in its versions YOLO-v5 [30], YOLO-v7 [31] and YOLO-v8 [32].
We trained and evaluated our models on a computer with these characteristics: AMD A12-9700P RADEON R7, 10 COMPUTE CORES 4C+6G at 2.50 GHz, 12 GB RAM, Windows 10 Home 64-bit operating system and x64 processor.
The development environment used was Google Colab with GPU accelerator type A100. Figure 1 shows the general architecture of YOLO, taking as reference the study presented by [33, 34]. The main components of YOLO are listed below:
– Backbone: The backbone is usually a convolutional neural network that extracts useful features from the input image [33].
– Neck: The neural network neck is used to extract features from images at different stages of the backbone. YOLOv4 make use of Spatial Pyramid Pooling (SPP) [35] and Path Aggregation Network (PAN) [36].
– Head: Is the final component of the object detector; this component is responsible for making the predictions from the features provided by the spine and neck [33].
In Figure 2, we show the methodology used for the detection of wildlife species in the Peruvian Amazon using transfer learning. It is composed of four phases, they are: Obtain images, images preprocessing, training models and testing and getting metrics. Each step of the proposed methodology is detailed below:
2.1 Image Obtaining
At this stage, we have searched for wild animals’ images by their scientific name. The images were collected from websites related to ecology studies and tourism marketing; such as Rainforest expeditions [37], Go2peru [38], Ararauna Tambopata [39].
Then, in order to download the images with high resolution, we have used the Fatkun Batch Download Image extension in its version 5.7.7 by the Google Chrome [40], as it was used in previous studies [41]. Finally, we have selected and filter manually only the images in jpg format.
2.2 Image Processing
This stage encompassed the process of image curation, organization and labeling. The curation of images was made according to the species to be identified, since the search yielded images related to the keyword. In Table 1, It is shown the total summary of images by species in the first dataset after selection.
Table 1 Number of images per wildlife species
| Species | Quantity |
| Ara ararauna | 232 |
| Ara chloropterus | 50 |
| Ara Macao | 52 |
| Opisthocomus hoazin | 100 |
| Pteronura brasiliensis | 110 |
| Saimiri sciureus | 109 |
| Total | 653 |
We have performed the images labeling manually. For this purpose, we have used the labelImg tool [42]. In Figure 3, we show an example of this labeling task in the specie Saimiri sciureus. As a result of this process, a textual file is generated and it fulfills the mission to designate each image.
Internally this file encompasses both the class to which the image belongs and the coordinates delimiting the bounding boxes containing the image. Finally, we have divided the dataset as follow:
We divided the dataset into 85% (556 images) for training, 10% (65 images) for validation and 5% (32 images) for testing. Figure 4 shows this division graphically.
In Figure 5 we show a summary of the distribution of classes in the datasets used to train, validate and test the species detection models based on the YOLO architecture.
2.3 Trining Model
During this phase we conducted out our experiments with the object detection algorithm YOLO in its versions: YOLOv5x6, YOLOv5l6, YOLOv7-W6, YOLOv7-E6, YOLOv8I and YOLOv8x. Within the file called ‘custom_data.yaml’ we have defined the configuration of the path to the training, validation and test images.
In the same file, we have additionally configurated the classes as follow: Scientific names: [Ara_ararauna, Ara_chloropterus, Ara_macao, Opisthocomus_hoazin, Pteronura_brasiliensis, Saimiri_sciureus].
2.4 Testing and Getting Metrics
We performed our tests with a video adapted from the public videos: "Vive como sueñas | Reserva Nacional Tambopata" from the Ministry of Environment [43, 44] and the video "Manu & Tambopata'' from the Antara-Peru travel agency [45]. Figure 6 and Figure 7, respectively, show a screenshot of these videos.
We have used the metrics: Precision, Recall, F1-Score and Mean Average Precision (mAP), and additionally the confusion matrix in order to evaluate the performance of the models. Subsequently we proceed to detail each of these metrics: Confusion matrix: It is an , dimension table where n represents the number of classes or objects to be detected.
This metric allows to evaluate the performance of a classification algorithm by counting the hits and mistakes in each one of the model classes. A confusion matrix for three classes is observed in Table 2, which can be extrapolated to object detection and classification problems with n classes.
Table 2 Confusion matrix for three classes
| Prediction | ||||
| True | A | B | C | FN |
| A | ||||
| B | ||||
| C | ||||
| FP | ||||
where:
FN represents false negatives.
FP represents false positives.
The true negatives (TN) for class:
A=
The true negatives (TN) for class:
B=
The true negatives (TN) for class:
C=
Precision: This metric, also known as positive predictive value (PPV) indicates the proportion of cases correctly identified as belonging to a specific class (e.g., class C) among all cases where the classifier claims to belong to that class.
In other words, accuracy answers the question: Considering that the classifier predicts that a sample belongs to class C, what is the probability that the sample actually belongs to class C? [46, 47]. Equation 1 illustrates the calculation of this metric:
where:
TP=True Positive.
FP=False Positive.
Recall: This metric, is also referred to as Sensitivity or True Positive Rate (TPR) measures the ratio of positive correctly identified positive cases (for our case study it represents the species to be identified) by the algorithm [48] Equation 2 shows the formula for calculating this metric:
F1-Score: It is defined as a harmonic mean of precision and recall. The F1 score reaches its best value at 1 and its worst value at 0. Equation 3 shows the formula for calculating this metric:
Average Precision (AP): This metric represents the relationship between precision and recall at different confidence thresholds, in addition to quantifying the ability of the detection model to discriminate between positive and negative classes. It is calculated from the Precision-Recall curve (PR Curve). Its value varies between 0 and 1, where an AP of 1 indicates perfect detection and AP of 0 indicates random detection [49, 51].
The mathematical operation for this calculation is shown in Equation 4:
where P and R refer to the precision and recall of the detection model that we detail in Equations (1) and (2), respectively. Mean average precision (mAP): We have calculated this metric by averaging the Average Precision (AP) values for all classes present in the dataset.
Its value fluctuates between o and 1, where 1 indicates perfect performance, i.e., all detections are correct and there are no false positives or false negatives. In Equation 5 we show the formula for the calculation of this metric:
3 Results and Discussions
Table 3, shows the configurations we have made for each YOLO version we used within our experimental framework, along with their corresponding training duration measured in minutes. Our selection process adhered to the guidelines outlined in the official documentation. According to this, we have chosen the two best pre-trained models per version [52]– [55]. We have made a particular exception for YOLOv7 because the YOLOv7-D6 and YOLOv7-E6E versions, which have a slightly better AP value, required a high computational cost for training, so we opted to use the YOLOv7-W6 and YOLOv7-E6 versions.
Table 3 Configuration and training times for models
| Model | Epochs | Batch | Input (resolution) | Training time in minutes |
| YOLOv5x6 | 70 | 16 | 640x640 | 57.94 |
| YOLOv5l6 | 70 | 16 | 640x640 | 30.71 |
| YOLOv7-W6 | 70 | 16 | 640x640 | 57.48 |
| YOLOv7-E6 | 70 | 16 | 640x640 | 60.36 |
| YOLOv8l | 70 | 16 | 640x640 | 37.14 |
| YOLOv8x | 70 | 16 | 640x640 | 62.20 |
Figure 8 presents the metrics for evaluating and monitoring of the selected model’s performance during training. The metrics are focused on the prediction accuracy of the object bounding boxes (box_loss) coordinates, error in the prediction of the classes of the detected objects (cls_loss), precision and recall.
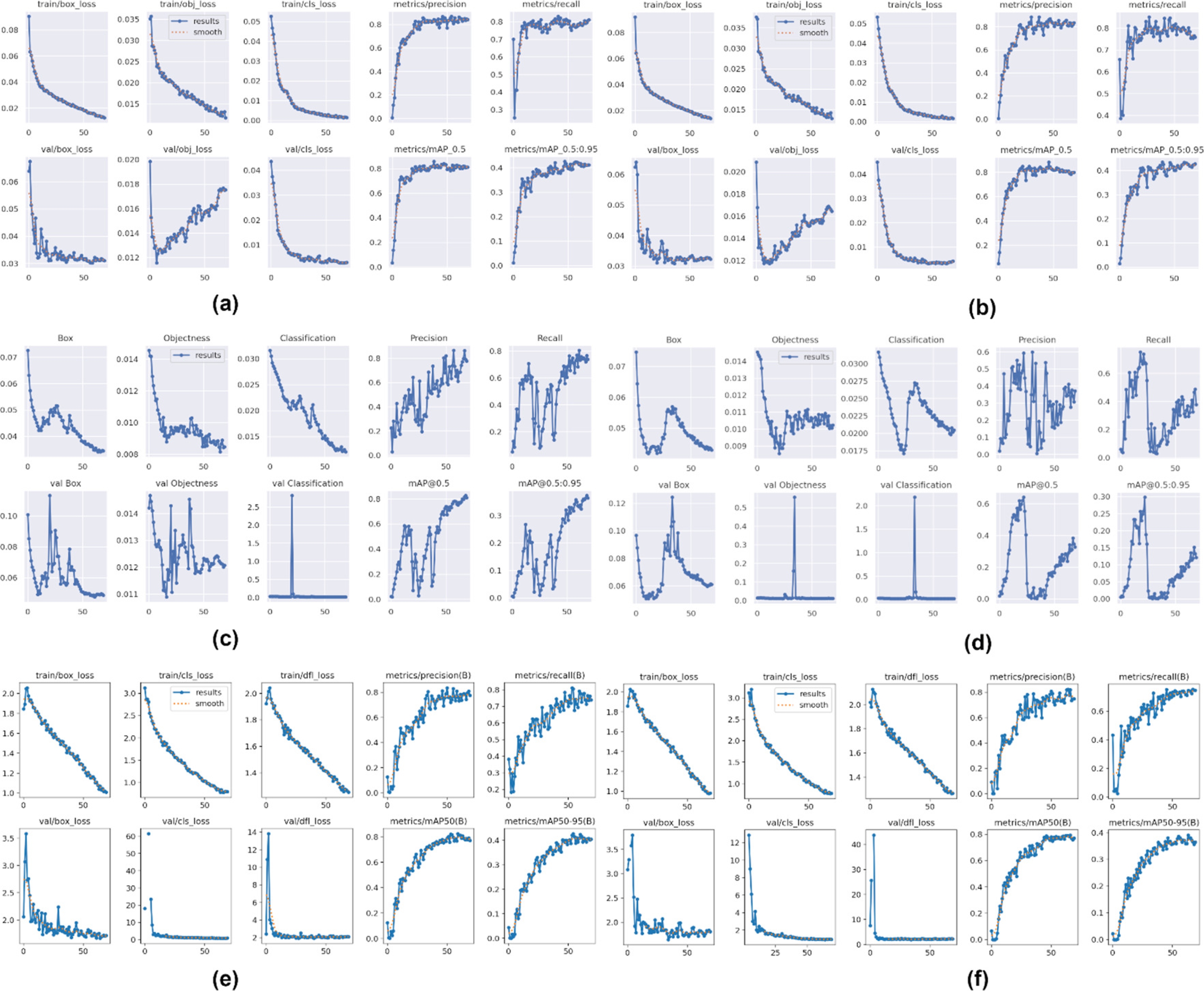
Fig. 8 Evaluation and performing metrics of the models during training. (a) YOLOv5x6 model, (b) YOLOv5l6 model, (c) YOLOv7-W6 model, (d) YOLOv7-E6 model, (e) YOLOv8l model and (f) YOLOv8x model
It is noted that the YOLOv5 (Figure 8a and 8b) and YOLOv8 (Figure 8e and 8f) models exhibit remarkable stability and out-standing performance in the task of accurate bounding box localization. These models demonstrate a consistent tendency to reduce the loss associated with accuracy, which holds important relevance in image object detection applications.
Specifically, in our domain study related to wildlife species identification, the results obtained by YOLOv5 and YOLOv8 show a superior ability to accurately localize the bounding boxes of the interest objects.
Moreover, during the validation stage, it is observed that these models maintain their stability, which confirms their robustness and their potential for practical applications in the field of computer vision.
With regard to the "cls_loss" metric, which reflects the discrepancy between model’s classification predictions and the actual labels of the object classes, it is graphically observed in Figure 8a and 8b that the YOLOv5 model manages to efficiently reduce this value down to epoch 50.
This suggests that as the model is trained its performance is better in the classification accuracy of wildlife species in the Peruvian Amazon.
Figure 9 shows the normalized confusion matrices during the training phase for the models utilized in our experiments. AA corresponds Ara ararauna species; AC, Ara cholopterus; AM, Ara macau; OH, Opisthocomus hoazin; PB, Pteronura brasiliensis and SS; Saimiri sciureus.
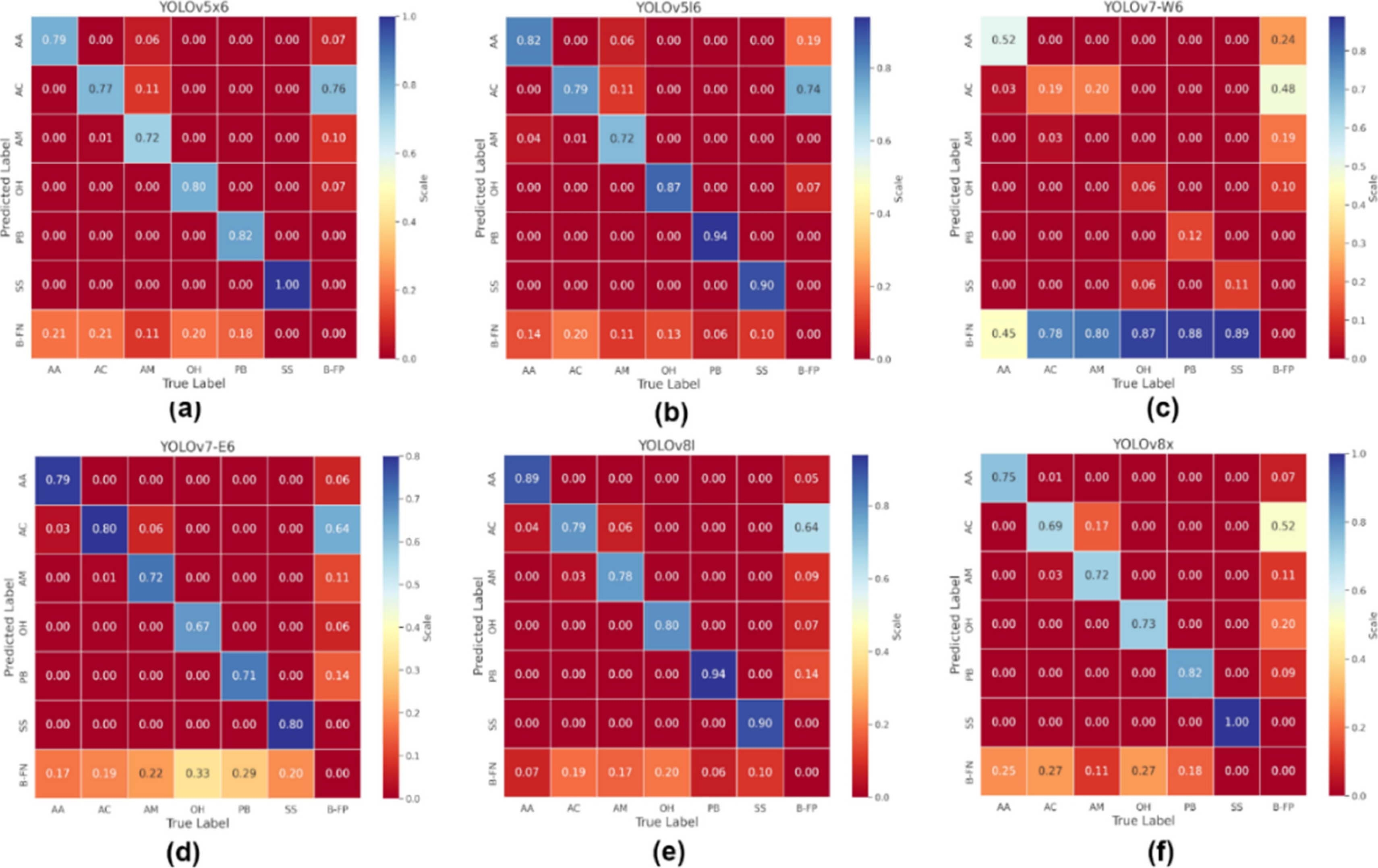
Fig. 9 Standard confusion matrices. (a) YOLOv5x6 model, (b) YOLOv5l6 model, (c) YOLOv7-W6 model, (d) YOLOv7-E6 model, (e) YOLOv8I model and (f) YOLOv8x model
In this graphic it is also observed that YOLOv8l y YOLOv5l6 models attain the highest values along the main diagonal. This diagonal represents instances in which the predicted labels by the aforementioned models align with the actual labels and suggests that in our dataset of Peruvian Amazon wildlife, the YOLOv8l and YOLOv5l6 models effectively identify the six species studied. Table 4 presents the results of the obtained metrics for each model in our experiments in detection of the six Peruvian Amazon wildlife species. The best values per species and metric are highlighted.
Table 4 Results obtained per evaluation metric
| Model | Class | Precision | Recall | F1-Score | mAP50 |
| YOLOv5x6 | All | 0.822 | 0.787 | 0.804 | 0.839 |
| Ara_ararauna | 0.842 | 0.786 | 0.813 | 0.857 | |
| Ara_chloropterus | 0.743 | 0.760 | 0.751 | 0.790 | |
| Ara_macao | 0.848 | 0.778 | 0.811 | 0.852 | |
| Opisthocomus_hoazin | 0.819 | 0.733 | 0.774 | 0.747 | |
| Pteronura_brasiliensis | 0.812 | 0.763 | 0.787 | 0.812 | |
| Saimiri_sciureus | 0.870 | 0.900 | 0.885 | 0.978 | |
| YOLOv5l6 | All | 0.861 | 0.847 | 0.854 | 0.881 |
| Ara_ararauna | 0.842 | 0.857 | 0.849 | 0.867 | |
| Ara_chloropterus | 0.727 | 0.745 | 0.736 | 0.816 | |
| Ara_macao | 0.781 | 0.722 | 0.750 | 0.759 | |
| Opisthocomus_hoazin | 0.864 | 0.849 | 0.856 | 0.871 | |
| Pteronura_brasiliensis | 0.951 | 0.941 | 0.946 | 0.980 | |
| Saimiri_sciureus | 1.000 | 0.967 | 0.983 | 0.995 | |
| YOLOv7-W6 | All | 0.371 | 0.378 | 0.374 | 0.327 |
| Ara_ararauna | 0.326 | 0.571 | 0.415 | 0.514 | |
| Ara_chloropterus | 0.373 | 0.480 | 0.420 | 0.349 | |
| Ara_macao | 0.080 | 0.218 | 0.117 | 0.065 | |
| Opisthocomus_hoazin | 0.240 | 0.267 | 0.253 | 0.225 | |
| Pteronura_brasiliensis | 0.493 | 0.229 | 0.313 | 0.327 | |
| Saimiri_sciureus | 0.714 | 0.500 | 0.588 | 0.483 | |
| YOLOv7-E6 | All | 0.776 | 0.739 | 0.757 | 0.808 |
| Ara_ararauna | 0.851 | 0.821 | 0.836 | 0.864 | |
| Ara_chloropterus | 0.700 | 0.779 | 0.737 | 0.765 | |
| Ara_macao | 0.753 | 0.722 | 0.737 | 0.792 | |
| Opisthocomus_hoazin | 0.832 | 0.662 | 0.737 | 0.791 | |
| Pteronura_brasiliensis | 0.613 | 0.647 | 0.630 | 0.681 | |
| Saimiri_sciureus | 0.909 | 0.800 | 0.851 | 0.952 | |
| YOLOv8I | All | 0.743 | 0.806 | 0.773 | 0.817 |
| Ara_ararauna | 0.828 | 0.893 | 0.859 | 0.910 | |
| Ara_chloropterus | 0.604 | 0.720 | 0.657 | 0.645 | |
| Ara_macao | 0.736 | 0.722 | 0.729 | 0.732 | |
| Opisthocomus_hoazin | 0.737 | 0.733 | 0.735 | 0.823 | |
| Pteronura_brasiliensis | 0.654 | 0.891 | 0.754 | 0.863 | |
| Saimiri_sciureus | 0.897 | 0.877 | 0.887 | 0.930 | |
| YOLOv8x | All | 0.819 | 0.745 | 0.780 | 0.790 |
| Ara_ararauna | 0.843 | 0.714 | 0.773 | 0.836 | |
| Ara_chloropterus | 0.711 | 0.624 | 0.665 | 0.698 | |
| Ara_macao | 0.820 | 0.758 | 0.788 | 0.795 | |
| Opisthocomus_hoazin | 0.784 | 0.726 | 0.754 | 0.694 | |
| Pteronura_brasiliensis | 0.856 | 0.765 | 0.808 | 0.820 | |
| Saimiri_sciureus | 0.898 | 0.885 | 0.891 | 0.898 |
It is important to stand out that the YOLOv5I6 model has demonstrated an outstanding performance by obtaining the highest values of Presicion, Recall, F1-Score and mAP50 for three of the six species analyzed: Opistho-comus hoazin, Pteronura brasiliensis and Saimiri sciureus.
The reference model demonstrated remarkable performance across evaluated metrics. It achieved a Precision rate of 86.4%, Recall of 84.9%, and an F1-Score of 85.6%, and a mAP50 of 87.1% for the first specie.
For the second one, the model exhibited exceptional Precision rate of 95.1%, paired with a Recall of 94.1%, an F1-Score of 94.6%, and an mAP50 of 98.0%.
The third specie yielded unprecedented results, attaining a perfect Precision of 100%, while sustaining a Recall of 96.7%, contributing to an impressive F1-Score of 98.3%. Remarkably, the mAP50 reached an astounding 99.5%. It's worth noting that this model consistently outperforms others, particularly evident in its remarkable mAP50 of 81.6% for the Ara chloropterus species.
Summarizing, our experiments have evidenced that the YOLOv5I6 model proves highly effective in specific species detection and it is notably excelling in the cases of Opisthocomus hoazin, Pteronura brasiliensis and Saimiri sciureus, such us in Ara chloropterus specie registering the highest mAP50 value.
On the other hand, the YOLOv5x6 model has also yielded remarkable results, particularly for the Ara macao species, in addition to obtaining higher values in the precision and F1 score metrics for Ara chloropterus. In Figure 10, we present a visual summary of the evaluated metrics for the six analyzed models. Our experiments highlight that the YOLOv5I6 model achieves the highest values in all of the evaluated metrics with a Precision rate of 86.1%, a recall of 84.7%, an F1-Score of 85.39% and an mAP50 of 88.1%.
Attained values are closely trailed by those reported by the YOLOv5x6 model. Notably, the overall averages of the metrics used in this study firmly indicate the YOLOv5I model as the optimal choice for our specific case.
These findings resonate with the conclusions drawn by [24], who explored animal detection and classification through camera trap images. Their evaluation of YOLOv5, FCOS, and Cascade_R-CNN_HRNet32 models yielded an impressive average Precision of 97.9%, along with an approximate mAP50 of 81.2% across all models.
Likewise, [25] introduced a framework for detecting four endangered mammal species Viverra tangalunga, Prionailurus javanensis sumatranus, Rusa alfredi, and Sus cebifrons in the forests of Negros Island using the YOLOv5 model.
Their effort resulted in an average mAP50 of 91% and a commendable Precision of 91%. We posit that the disparities in the metric values observed are attributed to the size discrepancy between the species examined in our research and those in the referenced studies.
Specifically, the species within our study context are comparably smaller than those encompassed in the aforementioned research. Our findings hold high significance for applications in wildlife monitoring and conservation.
They underscore the effectiveness of the YOLOv5I6 model in detecting wildlife species within the expanse of the Peruvian Amazon. Furthermore, these outcomes establish robust basis for forthcoming research endeavors and initiatives aimed at safeguarding the biodiversity of Amazonian regions.
Figure 11 provides an enlightening overview of numerical results of confidence in bounding boxes in the detection of wildlife species in the Peruvian Amazon. These results are screenshot captures obtained from the execution of our models on a video file in mp4 format (minute 1:00), derived from the videos "Vive como sueñas | Reserva Nacional Tambopata" [56] and "Manu & Tambopata" [57].

Fig. 11 Detection outcomes with confidence values within bounding boxes for models. (a) YOLOv5x6, (b) YOLOv5l6, (c) YOLOv7-W6, (d) YOLOv7-E6, (e) YOLOv8l and (f) YOLOv8x
It is noteworthy that the YOLOv7-E6 model is notable for its ability to identify the highest number of species in the video under analysis. However, a more detailed evaluation of the confidence associated with the object bounding boxes yields more accurate data.
We have observed that the YOLOv5I6 (b) model achieves a confidence of 83% for the Opisthocomus hoazin species, which represents the highest figure within this specific category. Concerning the detection of Ara arauna and Saimiri sciureus species, confidence levels of 90% and 75% are attained respectively.
It is worth noting that these values are remarkably close to those obtained by the YOLOv5x6 version. At the captured moment depicted in the figure, it is evident that both the YOLOv5I6 and YOLOv5x6 models failed to identify the Ara macao species.
However, it is important to note that the YOLOv7-E6, YOLOv8I and YOLOv8x models successfully detected this species at the same minute of capture. These findings provide a visual and tangible perspective on how the models perform in wildlife detection in the Peruvian Amazon.
These observations are crucial for understanding the applicability of the models in real world situations and highlight the significance of prediction confidence, as well as the selection of appropriate models based on detection objectives.
4 Conclusions
In this study we comprehensively assess the effectiveness of the YOLO algorithm in its versions YOLOv5x6, YOLOv5l6, YOLOv7-W6, YOLOv7-E6, YOLOv8I, and YOLOv8x.
Our evaluation centers on their suitability for detecting six wildlife species within the Peruvian Amazon, namely: Ara ararauna, Ara chloropterus, Ara macao, Opisthocomus hoazin, Pteronura brasiliensis, and Saimiri sciureus.
To build a robust foundation for our analysis, we curated a specialized dataset by sourcing images from ecological and tourism outlets, including Rainforest Expeditions, Go2Peru, and Ararauna Tambopata. The meticulous curation process, executed under the guidance of a wildlife specialist.
Our findings prominently showcase the prowess of the YOLOv5l6 model, which exhibits exceptional performance across all evaluated metrics. Notably, it achieves an impressive accuracy of 86.1%, a recall rate of 84.7%, an F1-Score of 85.39%, and a mean Average Precision (mAP) of 88.1%.
Remarkably, this model also boasts the shortest training duration at a mere 30.71 minutes among all models scrutinized. Furthermore, our experimental outcomes reveal a striking similarity between the achievements of the YOLOv5l6 model and those of the YOLOv5x6 model.
This convergence of results underscores the consistency and reliability of our evaluation methodology. These outcomes stand as promising and auspicious strides forward in fortifying initiatives aimed at identifying Amazonian wildlife species and vigilantly monitoring those that could potentially slip into states of vulnerability or endangerment.
By showcasing the potential of advanced algorithms, we not only demonstrate the power of technology but also emphasize the significance of collective efforts in safeguarding the rich biodiversity of the Amazon rainforest.
Data Availability Statement: We make our dataset and source code available to the scientific community at the following e-mail addressfn Additionally, you can watch the video used for the tests. There are also 6 videos that were generated for each model in the validation stage, these videos show the bounding boxes and their respective confidence valuefn.











 nueva página del texto (beta)
nueva página del texto (beta)










