Mesoamerica and South America, with genetically differentiable populations, but with a shared ancestor, are the center of origin and diversification of the common bean (Phaseolus vulgaris), with at least 8000 years of domestication (Rendón-Anaya et al., 2017; Bitocchi et al., 2017). Approximately 400 wild species of Phaseolus spp. with great morphological and physiological variability are estimated, of which five have been domesticated: P. vulgaris, P. lunatus, P. coccineus, P. acutifolius and P. polyanthus (De Ron and Santalla, 2013). Taxonomic and phylogenetic studies estimate 52 species in Mexico, of which 55% are endemic, with Jalisco, Durango and Oaxaca being the regions with the greatest diversity (Delgado and Gama, 2015). In Mexico, the crop has great historical agricultural, sociocultural, economic and nutritional relevance. Its diversity and productive adaptability are evidenced by more than 100 registered varieties (SADER and SNICS, 2022), highlighting Oti and black testa materials such as Negro Perla, Negro Jamapa, and others, for yield, grain quality, or resistance/tolerance to plant pathogens. Worldwide, Mexico is the sixth largest producer (FAOSTAT, 2022), and one of the main per capita consumers with an annual average of 10.4 kg. At the end of 2020, 1 086 733 t were reported, of which, 71% are concentrated in Zacatecas (41%), Sinaloa (13%), Nayarit (9%), Chiapas (6%), and Durango (4%) (SIAP, 2020). This production satisfies 90% of domestic demand, and 10% is imported from the USA, Canada and China.
The prolonged process of domestication and adaptation to diverse environments has detonated parasitic processes of co-evolution between Phaseolus spp. and different organisms, highlighting 36 species of phytopathogenic fungi e.g., Alternaria alternata, Pseudocercospora griseola, and Colletotrichum lindemuthianum (Berrouet-Vanegas et al., 2014); 26 viral organisms, e.g., Bean common mosaic virus, Bean golden yellow mosaic virus, and Bean common necrotic mosaic virus (Flores-Estévez et al., 2003); 22 nematode species, e.g., Dolichodorus heterocephalus, and Aphelenchoides ritzemabosi (Bird and Warner, 2018); nine oomycetes, e.g., Phytophthora nicotianae, and Pythium irregulare (Watanabe et al., 2007); and six bacterial species (e.g., Pseudomonas syringae pv. syringae, Curtobacterium flaccumfaciens, and Xanthomonas axonopodis pv. phaseoli) (Torres et al., 2009; Gent et al., 2005). Some of these organisms can be endemic, cause syndromes, or occur in mixed infections compromising yield and varietal stability (Pedroza-Sandoval et al., 2013; Lepe-Soltero et al., 2012; Mena and Velázquez, 2010; Estrada-Gómez et al., 2004; Flores-Estévez et al., 2003).
Seed, as the main sowing practice for P. vulgaris, represents one of the main risks for the dissemination of Potyvirus, Begomovirus and fungi such as Alternaria spp. (Subramanya, 2013). These organisms are widely distributed in Mexico, although with limited epidemiological studies. Systemic viral infections in beans, from infected seed, may cause greater productive detriment and loss of plant vigor due to chronic physiological alterations. Symptomatically, these can be expressed as poor plant development, dwarf, flower, and fruit abortion, and mosaics of yellow to golden tones, chlorosis, epinasty, distortion, and greening of the nervures on leaves (Rojas et al., 2018). In addition, 75% of bean pathogenic viruses are associated with insect vectors (aphids and whiteflies) increasing transmission and dispersal between plants and crop fields (Gilbertson et al., 2015). Among fungal problems, Alternaria spp. is not considered of high productive impact on beans despite its endemic status and to cause a typical blight with concentric necrotic spots affecting leaf tissue, pods and seeds with defoliation in advanced stages (Mena and Velázquez, 2010). The transmission of Alternaria spp. by seed represents an additional phytosanitary risk that should be investigated (Prasad and Ahir, 2013; Moraes and Menten, 2006).
In the bean crop, strategic in Mexico’s food security, systematic and comprehensive studies that articulate health, genetic, and production are limited. This approach would represent a paradigm shift that implies a regional, multidimensional and multi-pest vision to analyze the parasitic and epidemiological processes associated with production risks (Mora-Aguilera et al., 2021). The objective of this research was to develop etiological-epidemiological methodologies applicable to breeding programs in bean (P. vulgaris) with an integral productive and phytosanitary aim. For this purpose, 12 genotypes of P. vulgaris, exposed to natural infections of plant pathogens, were studied in two contrasting phenological events during the spring-summer 2020 season in a region of Mexican central highlands.
Materials and methods
Experimental unit. In May 2020, spring-summer season, a 5 000 m2 plot was selected at the Colegio de Postgraduados Campus Montecillo, Texcoco, Mexico State. Twelve Phaseolus vulgaris genotypes were sown in randomized blocks: Bayo Mecentral, Flor de Mayo, Tipo Flor de Mayo, Pinto Texcoco, Canario, Negro Mixteco, Negro Querétaro, Negro Perla, Vaquita Negro, Garrapato, OTI, and Segregante OTI (Figure 1A). Each genotype was established in 25 rows 13 m long with 80 cm, and 2 m spacing between furrows and blocks, respectively. A barrier of five maize (Zea mays) rows delimited the experimental area. A semi-technological agronomic management was implemented with conventional fertilization, hilling, weed control, and complementary irrigation. A HOBO u23 Pro v2 climate sensor was installed at the centroid of the experimental unit to measure relative humidity (RH) and temperature (°C), at 30 min intervals, between 1-June and 15-August 2020. Complementarily climatic data were obtained from the meteorological station of the Colegio de Postgraduados Campus Montecillo.

Figure 1 A) Experimental design of 12 Phaseolus vulgaris genotypes randomly distributed under field conditions. Aerial image captured on June 2, 2020, at 50 m from the centroid of the experimental unit using a Phantom 3 DJI® drone. Six-class logarithmic-diagrammatic scale to evaluate virus disease severity, estimated with percentage of mosaic-chlorosis-deformation leaf tissue in a 70 cm row-section (B), and to evaluate blight severity estimated with percentage of necrotic leaf area (C). The scale values apply to both diseases. LI, LS and PMC correspond to the lower, upper and midpoint of class in percentage units, respectively. Spring-Summer 2020 cycle.
Experimental variables and severity. Using App-Monitor® v1.1 Android®, available at PlayStore® (CP-LANREF, 2021), the coordinates, spatial location (state, municipality, and locality), crop name, plot extension, crop phenology, technification, irrigation, plantation density, and owner’s name were recorded in the characterization digital module. Epidemiological variables were evaluated in 13 plants/row in 5-6 rows/genotype by means of a 1 x 2 discontinuous systematic sampling. Two evaluations were made, one on flowering (6-June), and other on the fruit setting stage (10-August). Per plant, the virus symptoms severity, and leaf blight, the two infectious problems with the highest occurrence among genotypes, were evaluated. In addition, the presence or absence of Bemisia sp. and Aphis spp., as potential virus vectors, was reported.
Percentage of severity was evaluated using a 6-class logarithmic-diagramatic scale (González-Cruces et al., 2020). Class midpoint (CMP), lower limit (LL) and upper limit (UL) were calculated in 2-Log v2.0 employing the parameters of Number of classes = 6 and Y max = 65% (CP-LANREF, 2018. Unpublished). With virus symptoms, severity represented the percentage of mosaic-chlorosis-deformation tissue in a 70 cm row-section (Figure 1B); in blight, severity indicated the necrotic percentage of leaf area of the most infected leaflet/plant (Figure 1C). The severity scales were configured in App-Monitor®. A total of 12 assessors performed the measurements on each date. Data were stored in the App and subsequently exported in MS Excel for integration of the epidemiological data matrix and further system approach analysis (Mora-Aguilera et al., 2021).
Vigor Index. A coverage-plant vigor analysis was carried out during the flowering stage, assisted by 14 mpx RGB digital aerial images captured by a DJI® Phantom 3 drone between 15:00 and 17:00 h. The flights were performed in vertical trajectory from the centroid of the experimental unit (50 m height) and of each plot per genotype (3 and 5 m). The images were processed with Gimp® v2.10.20. By genotype, a Coverage Vigor Index (IV) = [(CF Gi )/(AT Gi )]*100 was obtained, where Gi is genotype-i, CF is coverage-plant area, and AT is total area (soil + coverage-plant).
Integrated Damage Index. In order to evaluate integrated health, with virus and blight severity data, and the Vigor Index as a correction factor, an Integrated Damage Index was calculated by genotype: (IDI) = [(3(CV Gi ) + 1 (AL Gi ))/260] + (1-IV Gi ), where Gi is genotype-i, CV percentage of virus disease severity, AL percentage of blight severity, and IV Coverage Vigor Index.
Sampling of plant material. In order to identify the agent(s) associated with blight, five samples were collected from the three genotypes with the highest incidence: Flor de Mayo, Pinto Texcoco, and Canario. Samples were collected targeting leaflets with concentric necrotic spots and leaf with 1 - 3 severity classes (0.01 - 20%) of the severity scale (Figure 1C). For virus symptoms, 20 and 18 trifoliate samples were collected with mosaic-epinasty and severe chlorosis-wrinkling symptoms, putatively associated with Potyvirus and Begomovirus, respectively. Sampling was representatively performed on the 12 genotypes.
Isolation and characterization of the blight-associated organism. A total of five symptomatic leaves of the genotype Flor de Mayo were cut into 10 5-mm squares from the marginal lesion zone and disinfested for 2 min with 1% sodium hypochlorite (NaClO), followed by three washes with sterile distilled water. Five mycelial growths were re-isolated and incubated in Petri dishes with Potato-Dextrose-Agar (PDA) medium (Difco®). After seven days, the sporulation pattern, conidiophores and conidia were reviewed using a stereo microscope (IROSCOPE YZ-6). With the hyphal tip technique, the isolates were purified on PDA and a mycelial growth characterization was performed. Mycelial color, morphology, and conidial morphometry were determined on 64 conidia/isolate. Fixed slides were made for compound microscopy (Velab VE-B2 of 10x and 40x) integrated to the Motic Images Plus v2.0 program. Identification was performed with taxonomic keys (Simmons, 2007; Barnet and Hunter, 1998). The characterization of isolates was documented photographically using a Canon EOS REBEL T6, 24.1 MP®.
Koch’s postulates. Twelve apparently healthy Flor de Mayo leaflets (4-5 cm wide and 5-6 cm long) were selected. They were disinfested with the previously described technique and dried in a laminar flow hood for 30 min. The 12 leaflets were placed individually in the center of Petri dishes, used as humid chambers, with an upper mesh and sterile absorbent paper below. Three treatments were carried out with three replicates and an absolute control. The treatments were inoculum of pure FPTA2h, FPTA3h, and FPTA4h isolates obtained from leaf blight and selected at random. Four discs with full mycelial growth (0.5 cm diameter)/isolate were distributed on the upper leaf blade. The absolute control consisted of leaflets without inoculation. The Petri dishes were incubated in a growth chamber at 25 °C and 90% relative humidity (RH) with a 12:12 h photoperiod for 7 d. Seven days after inoculation, the pathogenicity of each strain was evaluated by recording the presence or absence of typical blight symptoms. Six symptomatic leaves were selected, and the infective organism was re-isolated in Petri dishes with PDA. One colony per leaf was taken at random for morphological characterization.
Extraction of total nucleic acids. Extraction of total DNA from five, and total DNA and RNA from 5 and 37 samples was performed by the modified AP (SDS1%) method (Green and Sambrook, 2012). A total of 0.1 g of pure culture mycelium and leaf tissue was used for identification by partial genomic sequence of the fungus and putative viral agent(s), respectively. Optimal thresholds of nucleic acid concentration and purity were quantified with a NanoDrop spectrophotometer 2000 (Thermo Fisher Scientific 2000, USA).
Selection and in silico primers validation for genomic identification. Based on viral-like symptomatology in the field, infection of a member(s) of Potyvirus and/or Begomovirus genus was assumed. For genomic identification, three universal primer pairs were selected based on amplification on: 1) the nuclear inclusion coding region of protein B (NIb) in Potyvirus genomes; 2) the ORF BL1 of DNA-B in Begomovirus; and 3) the ITS region of the rDNA for eukaryotes (Table 1). The primers specificity associated with criteria 1 and 2 was assessed by local alignment using Blast® with genomic sequences registered at the National Center for Biotechnology Information (NCBI). For pathogens putatively associated with viral symptoms, a set of 22 nucleotide sequences specific for Bean golden yellow mosaic virus and Bean dwarf mosaic virus (Begomovirus), and Bean common mosaic virus (Potyvirus) reported in America for P. vulgaris were selected. The sequences were aligned with the respective primers using the Clustal W algorithm in MEGA 7.0.26 software.
Table 1 Name of the selected universal primer, sequence, and amplicon size for genomic identification of Potyvirus and Begomovirus species, and for eukaryotic microorganisms.
| Organismo | Iniciador | Secuencia | Tamaño de Amplicon (pb) | Cita |
|---|---|---|---|---|
| Potyvirus | NIb2F | 5' GTITGYGTIGAYGAYTTYAAYAA | 350 | Zheng et al., 2008 |
| NIb3R | 3' TCIACIACIGTIGAIGGYTGNCC | |||
| Begomovirus | PBL1v2040 | 5' CARTGRTCKATCTTCATACA | 500~650 | Rojas et al., 1993 |
| PCR1c | 3' CATATTTACRARWATGCCA | |||
| Hongo | ITS1 | 5' TCCGTAGGTGAACCTGCGG | ~500 | White et al., 1990 |
| ITS4 | 3' TCCTCCGCTTATTGATATGC |
Fungus molecular identification. Genomic identification of five fungal isolates was performed by amplifying the ITS region of the ribosomal DNA in a 25 µL final sample composed of: 1X PCR buffer, 1 mM MgCl2, 0.2 mM dNTPs mix, 400 nM of primers ITS1 and ITS4 (Table 1), 1 U of Taq DNA polymerase (Invitrogen), 40 ng of total DNA and nuclease-free water. Polymerase Chain Reaction (PCR) was performed on the T-100 thermal cycler (BioRad). A processing program was run consisting of initial denaturation at 95 °C for 5 min and 31 cycles with denaturation at 94 °C for 1 min, alignment at 55 °C for 1 min, extension at 72 °C for 90 s, and a final extension at 72 °C for 5 min. The amplified fragments were analyzed by 1.5% agarose gel electrophoresis stained with ethidium bromide and visualized with UV light in an imaging system (UVP, Biolmaging Systems, Epi Chemi II Darkroom).
Virus complex molecular identification. In 20 samples presumptively associated with Potyvirus symptoms, the synthesis of complementary DNA (cDNA) from total RNA was performed in two steps. In the first step, the reaction mixture with: 9.75 µL nuclease-free water, 500 nM per primer (NIb2F - NIb3R) and 2.5 µL total RNA, incubated at 85 °C for 3 min in a T100 thermocycler (Bio-Rad). In the second step, a mixture with: 2 mM mix dNTP’s, 1X buffer-RT, 100 U of reverse transcriptase enzyme (M-MLV-RT) and 10 U of ribonuclease inhibitor (RNAsin), all from Promega Corp. USA. The total reaction volume was 20 µL incubated at 44 °C for 60 min, followed by inactivation of the enzyme at 92 °C for 10 min. RT-PCR was performed in a final volume of 25 µL consisting of: 12.5 µL of Gotaq® G2 Hot star mix, 500 nM of NIb2F - NIb3R primers (Table 1) and 2.5 µL of cDNA. The final volume was completed with nuclease-free water. Using the T-100 thermal cycler (BioRad), a program was run consisting of: initial denaturation at 95 °C for 2 min, 35 cycles with denaturation at 95 °C for 45 s, alignment at 45 °C for 45 s, extension at 72 °C for 45 s and a final extension at 72 °C for 5 min.
For 18 putative Begomovirus samples, the PCR reaction mixture had a final volume of 25 µL containing: 1X GoTaq® buffer, 0.2 mM dNTP’s mix, 300 nM of the primers PBL1v2040 - PCR1c (Table 1), 0.5 U of GoTaq® G2 and 2 µL of DNA. The final volume was volumetrically gauged with nuclease-free water. The thermocycling program executed was initial denaturation at 95 °C for 3 min, 30 cycles with denaturation at 94 °C for 30 s, alignment at 50 °C for 30 s, extension at 72 °C for 45 s, and a final extension at 72 °C for 5 min. The amplified fragments of samples presumptive to Potyvirus and Begomovirus were analyzed by electrophoresis with the methodology analogous to the identification of the fungus.
Sequencing and bioinformatics analysis. A total of 23 samples that amplified by PCR were sent for nucleotide sequencing (Macrogen® Seoul, Korea). With the sequences and reference sequences for each organism group, bioinformatic analyses were performed including: 1) Generation of the consensus sequence with the SeqAssem v07 program; 2) Local alignment with Blast-n® at NCBI; 3) Evaluation of the percentage of homology and coverage with the amplified genomic region; and 4) Phylogenetic reconstruction in MEGA v7.0.26 using maximum likelihood and/or maximum parsimony as clustering methods.
Spatial analysis. The intra- and inter-plot geospatial analysis was performed with Golden Surfer® v10. The data matrix by phenological event and genotype was structured with x = row, y = plant spacing (fungus) or 70 cm row-section (virus), and z = percentage of severity of virus symptoms or blight. Geostatistical analysis was performed with the kriging method represented in two-dimensional maps and contours. Spatial dependence and autocorrelation were calculated with omnidirectional variograms fitted to a spherical model. By genotype, the graphic indicators nugget (n), σ2-partial (σ2-p), and sill of σ2 (σ2-s) were obtained to determine the level of continuous spatial dependence.
Abiotic damage associated with fungal infection. Due to the distribution of blight in the field, predominantly basal on the plant, and record of a hail event at the early crop stage, a simulated hail (G), wind (V) and soil friction (S) damage trial was conducted on leaf tissue of P. vulgaris to evaluate its implication in the infection process and blight development under greenhouse conditions. The trial was stablished with a total of nine asymptomatic Flor de Mayo leaflets, distributed in three replicates per treatment. The hail effect was simulated by impacting five frozen hydrogel spheres of 0.5 cm diameter at 10 cm from each leaflet. Wind simulation was performed by friction between leaflets by means of air generated with a fan (Taurus rush® 20 inches) at 1300 rpm, during 5 min. For the Soil effect, a total of 2 kg of soil was gradually dispersed over the leaf tissue. Absolute controls were asymptomatic leaflets, without exposure to damage and sprayed with sterile distilled water.
Immediately after the simulation, the upper blade of each leaflet was inoculated by spraying 20000 (C1), 39000 (C2) and 49000 (C3) conidia per mL-1 of the FPTA2h isolate. Each leaflet was placed in 2 mL tubes immersing the peduncle in sterile distilled water to maintain turgor during the experiment. This material was placed inside polypropylene bags to avoid contamination. From symptomatic leaves, the organism was re-isolated in Petri dishes with PDA eight days after inoculation. Six preparations of direct lesion structures were made for identification under compound microscopy (10x and 40x Velab VE-B2) using the previously cited taxonomic keys. Additionally, a digital collection of 12 images of damaged leaf tissue was generated to quantify the percentage of severity by processing with GIMP 2.10®. The results were analyzed in SAS v9.4 software using ANOVA in a BCA design and the PROC GLM procedure with the combination of simulated damage type (G, V, S, T) and concentration (C i ) as treatments.
Statistical analysis. An epidemiological matrix with 859 observations, 22 variables, and 18898 total metadata was integrated with MS Excel for statistical analysis in Statistical Analysis Software (SAS®) v9.4. Viral symptom severity, blight, Integrated Damage Index, and Vigor Index were used as response variables, whereas genotype and phenological event were experimental factors. Descriptive analyses were performed by variable and phenological events with PROC UNIVARIATE for normality tests through Shapiro-Wilk (p < 0.05). Analysis of variance (ANOVA) for a completely randomized design (CRD) with nested replications was performed with PROC GLM and Tukey mean comparison (α = 0.05). Comparison between phenological events was performed with t-Student (α = 0.05).
Results and discussion
Morphological and molecular fungus identification. The FTFMA1h, FPTA2h, FPTA3h, FPTA4h, and FPTA5h fungus isolates were characterized by black mycelial growth with 5 to 8 conidia-chain per conidiophore (Figure 2A). A total of 64 mature conidia of FPTA2h isolate were morphometrically measured, showing an average length and width of 7.53 µm (range 3.8 - 12.3 µm) and 3.23 µm (range 2 - 4.6 µm), respectively. Conidium had 3 - 5 transversal, and 1-2 longitudinal septa (Figure 2A). Based on these characteristics, the fungus was identified as Alternaria alternata (Berrouet et al., 2014; Simmons, 2007; Barnet and Hunter, 1998).
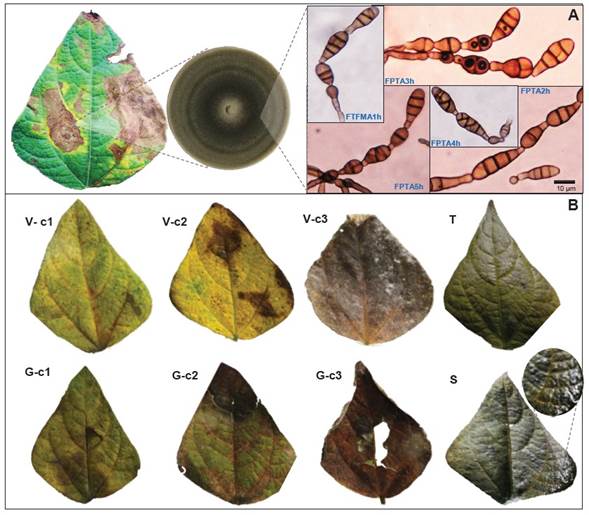
Figure 2 A. Alternaria alternata identification by symptom Association - Culturing - Identification based on morphometry of dyctiospores in chain, short pedicels, conidiophores (see description in text). B. Simulation of wind (V), hail (G), and soil friction (S) effect damage on Alternaria alternata infection under controlled conditions. c1 = 20000, c2 = 39000, c3 = 49000 conidia mL-1 FPTA2h isolate; T = Control. Except for S, which exhibited a bacterial-type biofilm, mechanical damage was favorable for fungus infection.
A. alternata was confirmed by molecular amplification of optimal fragments 500-600 bp ITS region. Threshold concentrations of 127.7 - 482.3 ng μL-1 and purity 1.88 - 2.18 nm were obtained. Five sequences were aligned at NCBI and confirmed the taxonomic identification of A. alternata with 99 - 100% homology regarding the A. alternata accession AF347031.1. The sequences were registered in GenBank with accession numbers OL229866, OL229867, OL229868, OL229869, and OL229870. The phylogenetic analysis, using the maximum likelihood method (i=1000) for ITS region and 100% node support, clustered all five sequences with the accession AF347031.1. The reference sequences of Alternaria alternantherae (KC584179.1) and Stemphylium amaranthi (KU850503.1) were clearly separated from the A. alternata node (Figure 3A). However, since A. alternata (Alternaria sect. alternaria) is an evolutionary diverse group, with morphological and molecular ITS attributes potentially overlapping among species, complementary studies may be necessary (Armitage et al., 2020).
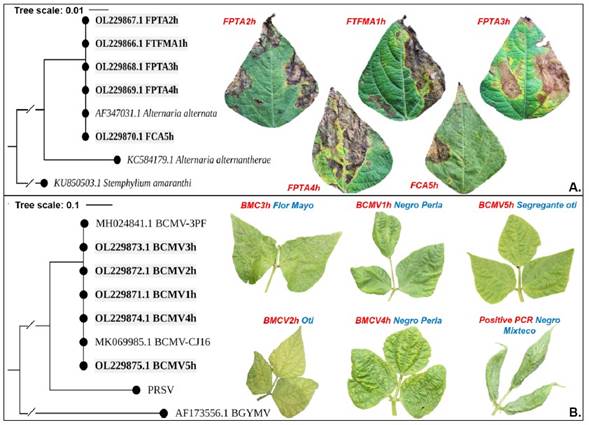
Figure 3 Bioinformatic analysis through amplified sequences of universal genomic regions and maximum likelihood with 1000 Bootstrap repeats. A) A. alternata identification through ITS1 and ITS4, HKY+G model with 99% homology with respect to GenBank isolate AF347031.1. B) Bean common mosaic virus identification through NIb2F and NIb3R, Tamura-Nei model with 99% homology with two accessions of this virus from GenBank. Bold sequences correspond to samples obtained in this work.
Alternaria alternata pathogenicity tests. Pathogenicity of FPTA2h, FPTA3h, and FPTA4h A. alternata isolates was confirmed on leaf tissue of P. vulgaris genotype Tipo Flor de Mayo. The control leaflets did not reproduced symptoms. On average, seven days after inoculation, 100% of leaflets showed symptoms as dark-brown or black necrotic lesions accompanied by chlorotic extending halos. Colonial and conidial morphometry adheres to the taxonomic characteristics of Alternaria alternata in all leaflets infected (Simmons, 2007; Barnet and Hunter, 1998), confirming Koch’s postulates.
Molecular diagnosis and prevalence of Potyvirus and Begomovirus. RT-PCR detected a 100% prevalence of one member of the Potyvirus genus in the samples analyzed (20/20). The 350-bp amplicon for universal primers NIb2F and NIb3R was amplified in all 12 genotypes of P. vulgaris samples. The concentration and purity thresholds ranged from 127.7 - 482.3 ng μL-1 and 1.88 - 2.18 nm, respectively. Sequencing of five representative samples identified Bean common mosaic virus (BCMV) specie at 99 - 100% homology with the NCBI accessions MH024841.1 BCMV-3PF and MK069985.1 BCMV-CJ16 of this virus. The phylogenetic construction using Maximum Likelihood, reached 100% node support (Figure 3B). Reference sequences of Papaya ringspot virus (PRSV), specie of the Potyvirus genus, and Bean golden yellow mosaic virus (BGYMV), belonging to Begomovirus genus, were separated of the main clade. BCMV confirmatory sequences in this paper were recorded in GenBank with accession numbers OL229871.1, OL229872.1, OL229873.1, OL229874.1, and OL229875.1.
The high incidence of this virus agreed with the 90.2 - 100% reports for Africa (Mwaipopo et al., 2018), although it exceeded that reported previously by Lepe-Soltero et al. (2012) in Sonora and Nayarit (6 - 78%). In this study, BCMV symptoms were mosaic (100%), epinasty (60%), and leaf distortion (39%), which were apparently influenced by the genotype (Figure 3B). The BCMNV was not detected, a Potyvirus closely related to BCMV, previously reported with higher prevalence in five Mexican states, State of Mexico not included, occurring in single infection or mixed with BCMV (Lepe-Soltero et al., 2012). Even though necrosis symptoms on secondary veins, typical of BCMNV, were not detected in this research, future studies may require specific primers, in addition to a universal sequence, to ensure the diagnosis.
The 18 samples with severe leaf yellowing suspected of some member of Begomovirus genus were not amplified with PBL1v2040 and PCR1c universal primers, despite the presence, although in low density of B. tabaci in over 80% of 12 genotypes. This insect represents a potential vector of this virus group. However, this finding was unexpected considering that Bean golden yellow mosaic virus (BGYMV) and Bean golden mosaic virus (BGMV) have been widely reported for Central America and the Caribbean (Rojas et al., 2018; 1993), although strongly associated with regional outbreaks of Bemisia tabaci in the 1990s (Gilbertson et al., 2005). In Brazil, BGMV was reported infecting seven bean cultivars, though at different viral concentrations (De Freitas-Vanzo et al., 2021).
Bean common mosaic virus severity in 12 bean genotypes. The epidemiological matrix for comparative analysis among genotypes regarding A. alternata and BCMV severity, and spatial spread, included 22 variables, 859 observations, and 18898 metadata. In flowering phase, BCMV plant severity varied between 9 and 50% (classes 3 - 5) with high genotype-dependent variability. Vaquita Negro (50.06 ± 13.6), Canario (48.7 ± 15.4), and Segregante Oti (40.14 ± 21.9) exhibited higher symptom expressions severity classes 4 - 5 compared to other genotypes (Tukey, p = 0.05) (Table 2). On the contrary, Bayo Mecentral (21.2 ± 8.1), Tipo Flor de Mayo (18.2 ± 14.8) and Negro Perla (9.6 ± 6.9) exhibited relatively lower severity (classes 3 - 4) (Tukey, p = 0.05) (Table 2). At fruit stage, severity range increased to 6.7 - 58.1% (classes 3 - 6). Vaquita Negro (58.1 ± 4.8) and Canario (37.3 ± 20.3) were statistically the most susceptible genotypes with severity classes 4 - 5 (Tukey, p = 0.05), whereas Garrapato (38.5 ± 18.5) which increased in severity by 14%, was statistically different (t-test, p < 0.0001) respect to the flowering stage.
Negro Perla (6.7 ± 8.8) was the lowest susceptible genotype at flowering and fruit stage without statistical differences (t-test, p = 0.12) (Table 2). However, the trend was a reduction between 2.9 - 19.9% severity, attributed to vegetative growth, previously documented in Kenya for pre- and post-rainfall experiments, where severity decreased up to 12.6% in ten P. vulgaris cultivars (Mangeni et al., 2020). In this research, eight genotypes showed the decreasing condition, of which six were statistically different (t-test, p = 0.0001). Severity for Flor de Mayo (- 6.4%) and Negro Perla (- 2.9%) was no statistical different in flowering and fruit stage (t-test, p = 0.12) (Table 2). Among the most susceptible genotypes, those with statistically significant increases (6.4 - 8.8%) (t-test, p = 0.021 - 0.042) were Vaquita Negro (58.1 ± 4.8) and Tipo Flor de Mayo (27 ± 22) (Table 2).
Table 2 BCMV and A. alternata average severity percentage in 12 bean genotypes (P. vulgaris) on flowering and fruit stage under field conditions. Spring-Summer 2020 season.
| Genotipo y | Bean common mosaic virus | Alternaria alternata | ||||
|---|---|---|---|---|---|---|
| Floración | Fructificación | t-test x | Floración | Fructificación | t-test x | |
| Vaquita Negro | 50.0 ± 13.6 a | 58.1 ± 4.8 a | 0.021 | 10.6 ± 10.6 b | 11.9 ± 6.0 b | 0.581 |
| Canario | 48.7 ± 15.4 a | 37.3 ± 20.3 abc | 0.000 | 7.0 ± 7.5 b | 42.7 ± 18.2 a | 0.000 |
| Segregante Oti | 40.1 ± 21.9 ab | 31.0 ± 16.4 abc | 0.039 | 6.8 ± 9.4 b | 8.6 ± 6.6 b | 0.424 |
| Flor de Mayo | 36.0 ± 15.4 abc | 29.8 ± 18.8 abc | 0.123 | 11.4 ± 10.1 b | 11.9 ± 9.2 b | 0.870 |
| Negro Mixteco | 31.5 ± 15.8 abc | 12.1 ± 12.3 bc | 0.000 | 16.7 ± 10.3 ab | 7.5 ± 4.2 b | 0.001 |
| Negro Querétaro | 29.9 ± 8.1 abc | 15.2 ± 12.4 bc | 0.000 | 15.3 ± 11.9 ab | 10.0 ± 10.8 b | 0.119 |
| Garrapato | 24.5 ± 19.1 abc | 38.5 ± 18.5 ab | 0.000 | 5.3 ± 4.3 b | 20.2 ± 8.6 b | 0.000 |
| Pinto Texcoco | 23.4 ± 10.5 abc | 16.0 ± 10.0 bc | 0.051 | 33.6 ± 8.1 a | 9.5 ± 4.8 b | 0.000 |
| Oti | 23.3 ± 21.0 abc | 29.7 ± 17.8 abc | 0.102 | 5.6 ± 7.4 b | 15.2 ± 7.4 b | 0.000 |
| Bayo Mecentral | 21.2 ± 8.1 abc | 9.5 ± 4.2 bc | 0.000 | 11.8 ± 7.4 b | 12.3 ± 3.4 b | 0.865 |
| Tipo Flor de Mayo | 18.2 ± 14.8 bc | 27.0 ± 22 bc | 0.042 | 9.9 ± 11.8 b | 41.4 ± 13 a | 0.000 |
| Negro Perla | 9.6 ± 6.9 c | 6.7 ± 8.8 c | 0.121 | 10.0 ± 10.6 b | 6.2 ± 5.8 b | 0.111 |
x Student’s t-test (α = 0.05) comparing mean percent BCMV severity in plant canopy and A. alternata foliar severity between flowering and fruiting. Treatments with at least a common letter are statistically equal ANOVA/Tukey p = 0.05. Bold numbers represent the genotypes with the maximum and minimum severity values of BCMV and A. alternata. Signs ± followed by a number represents the intra-genotype standard deviation (SD) of mean percent severity.
Overall, varietal studies of bean virus severity (Potyvirus, Begomovirus, or another genus) use qualitative leaf-level scales (e.g., 0 = no symptoms, 1 = mild symptoms, 2 = moderate symptoms, and 3 = severe distortion, leaf or stem malformation, and stunting). Using this scale, Mangeni and coworkers (2020) reported a 2.3 average severity of BCMV, which could be comparative to classes 4 - 5 of the severity scale developed for this study, found in Vaquita Negro, Canario, Garrapato, or Segregate Oti (Table 2). Murere and coworkers (2018) studying 16 bean genotypes reported BCMV prevalence in 29.4 - 42.78% cultivars with a regional average severity of 1.1 - 1.67. Susceptible genotypes exhibited moderate and severe symptoms (classes 2 - 3).
Regarding disease intensity caused by Begomoviruses (i.e., CuLCrV and SiGMFV) in Phaseolus sp., severity has been reported between 5 - 50% in 20 genotypes considered of moderate resistance, and over 60% for 21 varieties typified as susceptible (Agarwal et al., 2021). These findings show the high prevalence and severity of BCMV and other virus species on beans, suggesting the need of updating the viruses status in Mexican germplasm stocks and the productive implications (Flores-Estévez et al., 2003; Pedroza-Sandoval et al., 2013; Lepe-Soltero et al., 2012). The use of a 70 cm row-section plant-coverage as scale proposed in this work allowed discriminating the viral infectious effect and therefore constitutes a methodological alternative for intensive field studies with respect to plant-level scales under the rationale of confounding canopy in high density plantings.
Alternaria alternata severity in 12 bean genotypes. Among the 12 bean genotypes studied, A. alternata severity was highly variable. In the flowering stage, leaf severity ranged from 7 - 33.6% (classes 3 - 4). Pinto Texcoco was the most susceptible genotype (33.6 ± 8.1), followed by Negro Mixteco (16.7 ± 10.3) and Negro Querétaro (15.3 ± 11.9). However, these genotypes were no statistically different (Tukey, p = 0.05) due to intra- and inter-genotype variability (Table 2). Garrapato (5.3 ± 4.3), Oti (5.6 ± 7.4), and Segregante Oti (6.8 ± 9.4), belonging to the same lineage (Estrada-Gómez et al., 2004), exhibited tolerance to the fungus. These materials were equal statistically (Tukey, p = 0.05) respect to the other genotypes except for Pinto Texcoco (Table 2). This lineage was reported as highly or moderately resistant to foliar and root fungal infections such as Colleotrichum lindemuthianum, Sclerotinia sp., Uromyces appendiculathus var. appendiculatus and Rhizoctonia solani (Estrada-Gómez et al., 2004).
At the fruit stage, severity maintained its variability. Nevertheless, Canario (42.7 ± 18.2) and Tipo Flor de Mayo (41.4 ± 13) were the most susceptible genotypes showing significant increases compared to the flowering stage with 35.7% and 31.5% (t-test, p < 0.0001), respectively (Table 2). This increase is explained by greater plant longevity, favoring leaf growth and prolongation of infection cycles. The genotypes Negro Perla (6.2 ± 5.8), Negro Mixteco (7.5 ± 4.2), Segregante Oti (8.6 ± 6.6), Pinto Texcoco (9.5 ± 4.8), and Negro Querétaro (10 ± 10.8) exhibited lower severity than 10% (Classes 1 - 3), and a significant decrease severity, from 3.8 - 24.1%, respect to the flowering stage (t-test, p < 0.01) (Table 2). Pinto Texcoco exhibited the highest severity reduction from 33.6% to 9.5%, followed by Negro Mixteco decreased by 16.7%. Analogous to BCMV, the severity decrease in these genotypes was associated with tissue senescence and consequently inoculum loss. Bayo Mecentral, Flor de Mayo, Vaquita Negro, and Segregante Oti were the most stable genotypes, with moderate severity of 6.8 - 11.8% in the flowering stage, and not significant increases, between 0.4 - 1.8%, in the fruit stage (t-test, p = 0.42 - 0.87) (Table 2).
Several Alternaria spp. species has been reported associated with beans and other crops. i.e., A. solani in some species of the Solanaceae family can be highly restrictive for production (Thomma, 2003). Alternaria alternata (= A. tenuis) was reported as a pathogenic agent on Phaseolus spp. at least since the early 19th century (Thomma, 2003; O’Donnell and Dickinson, 1980). Nevertheless, epidemiological studies of A. alternata applied to P. vulgaris germplasm assessment were not found. This report is the first in Mexico with this approach.
Integrated Damage Index (IDI) and Vigor Index (IV). Comprehensive health of P. vulgaris genotypes regarding BCMV and A. alternata, estimated by IDI, ranged from 0 - 1.54. The IDI was higher at flowering stage between 0.37 - 1.51, being Oti (0.37) and Negro Perla (0.43) significantly more tolerant genotypes, whereas Vaquita Negro (1.51) and Segregante Oti (1.05) were the most susceptible to BCMV and A. alternata combined effect (Tukey, p = 0.05) (Table 3). In fruit phase, the IDI decreased considerably to ranges of 0.03 - 0.63. Once again, Negro Perla (0.03) and Oti (0.03) were statistically the most tolerant, and Vaquita Negro (0.63) was the most susceptible (Tukey, p = 0.05) (Table 3).
Table 3 Vigor Index (IV) in flowering phase and Integrated Damage Index (IDI) in flowering and fruit stages assessed in 12 varieties of bean (P. vulgaris) under field conditions. Spring-Summer 2020 season.
| Variedad y | IV x | IDI y Floración | IDI z Fructificación |
|---|---|---|---|
| Oti | 0.91 a | 0.37 f | 0.03 c |
| Negro Perla | 0.71 b | 0.43 ef | 0.03 c |
| Canario | 0.68 c | 0.90 bcd | 0.18 b |
| Flor de Mayo | 0.66 d | 0.79 bcd | 0.13 bc |
| Garrapato | 0.64 e | 0.66 def | 0.18 b |
| Pinto Texcoco | 0.61 f | 0.78 bcd | 0.08 bc |
| Tipo Flor de Mayo | 0.53 g | 0.71 cde | 0.21 b |
| Negro Querétaro | 0.48 h | 0.91 bcd | 0.10 bc |
| Bayo Mecentral | 0.47 hi | 0.81 bcd | 0.08 bc |
| Negro Mixteco | 0.45 ij | 0.97 bc | 0.09 bc |
| Segregante Oti | 0.43 j | 1.05 b | 0.22 b |
| Vaquita Negro | 0.10 k | 1.51 a | 0.63 a |
x Treatments with at least one common letter are statistically equal by Tukey p = 0.05. x Tratamientos con al menos una letra en común son estadísticamente iguales por Tukey p = 0.05.y Genotypes and IV and IDI values in bold represent maximum and minimum contrasts. High IV and low IDI values indicate higher tolerance to BCMV and A. alternata.
The Vigor Index (IV) by genotype ranged from 0.1 - 0.91. This index was inversely proportional to IDI, evidencing a strongly detrimental physiological effect of the BCMV and A. alternata combination. Overall, IV showed statistical differences at least in 8/12 genotypes (Tukey, p = 0.05). Oti (0.91) had significantly higher coverage-plant vigor (Tukey, p = 0.05). Interestingly, Garrapato, from whom Oti was derived, and was selected for Mexico State Valley due to high productivity and resistance to plant pathogenic fungi (Estrada-Gómez et al., 2004), exhibited moderate IV = 0.64 (Table 3), and moderate susceptibility with 38% maximum damage by BCMV and 20.2% A. alternata (Table 2). By contrast, Vaquita Negro (0.1) showed the lowest vigor, indicating high susceptibility, particularly to BCMV. Negro Querétaro, Bayo Mecentral, Negro Mixteco and Segregante Oti were similar genotypes in IV, ranged from 0.43 - 0.48 (Tukey, p = 0.05) (Table 3).
This research supports Oti and Negro Perla as the genotypes with the highest tolerance to BCMV and A. alternata, attributed to intrinsic genotype-environment characteristics. However, it is necessary to analyze lineages and parental lines to optimize this type of studies in comprehensive breeding programs that incorporate phytosanitary factors in addition to the phenotypic approach associated with yield components.
Spatial analysis of severity at genotype level. Geostatistical kriging analysis and omnidirectional variograms of Bean common mosaic virus and A. alternata exhibited random intra-genotype dispersal patterns of random foci, aggregates, and uniformity depending on susceptibility / tolerance and phenological phase. Aggregation intensity agreed with severity intensity / genotype, IDI, and IV results discussed in previous sections (Table 2, 3; Figure 1, 4).
The Vaquita Negro block, the most susceptible genotype to BCMV, was characterized by high-intensity dispersion (class 5, 58.2%) at fruit stage (Figure 4A), without spatial dependence of severity indicating uniform virus infection distribution through the parameters’ nugget[n]= 209, σ2-parcial[σ 2 -p] ≈ 0, σ2-sill[σ 2 -s] = 233.7 (Figure 4B- B11 ). On the contrary, Bayo Mecentral, although had a uniform distribution (n= 40.9, σ 2 -p ≈ 0, σ 2 -s = 70.5) was associated with low severity (class 3, 9.6%) (Figure 4A, 4B- B1 ).
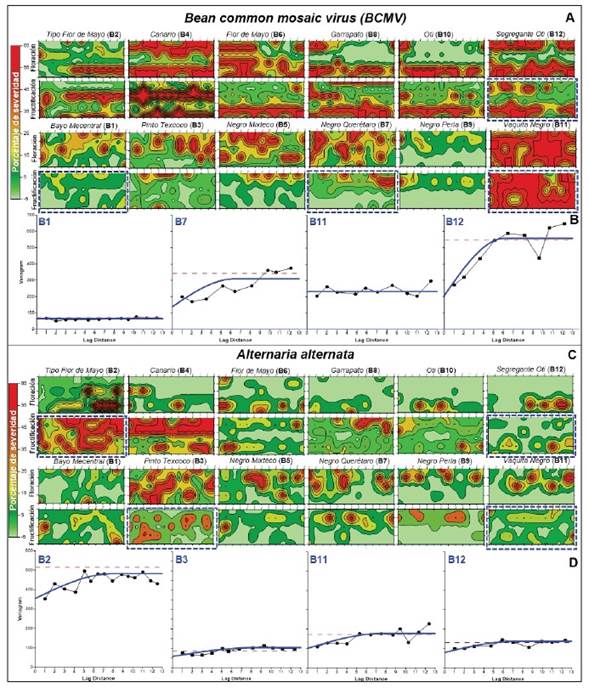
Figure 4 Geostatistical kriging analysis of average severity of A) Bean common mosaic virus and C) Alternaria alternata assessed in flowering (6 June, 2020) and fruiting (10 August, 2020) phase for 12 Phaseolus vulgaris genotypes. Percentage severity is represented in green-red colorimetry according to assessment scale classes (Figure 1). Omnidirectional semivariograms by spherical method for contrasting genotype block, highlighted with blue dashes boxes, for B) BCMV (B1, B7, B11, and B12) and D) A. alternata (B2, B3, B11, and B12) in fruiting phase. The X-axis represents lag -distance of 70 cm row-sections (BCMV), -plant (A. alternata), and Y severity variance (σ2).
Segregante Oti represented genotypes with spatial dependence in discontinuous aggregate foci of 4-8 distances of 70 cm row-sections (n= 207.4, σ 2 -p = 338.6, σ 2 -s = 546) (Figure 4A, 4B- B12 ), analogous to Tipo Flor de Mayo and Negro Mixteco, or block-edge effect, as Canario, Flor de Mayo, Garrapato, and Oti genotype (Figure 4A, 4B). Other group included Negro Querétaro with random foci (n= 136, σ 2 -p > 207.8, σ 2 -s = 343.8) (Figure 4A, 4B- B7 ), similar to Bayo Mecentral, Pinto Texcoco and Negro Perla, this last one significantly tolerant for both phenological stages (Figure 4A, 4B). Uniform dispersal pattern associated with BCMV, without statistical differences between phenological events (data not shown), suggests a seed-borne transmission effect, which is well stablished for this virus (Worral et al., 2015; Subramanya, 2013). However, a vector(s) may also be involved due to random foci and edge-block effect. Myzus persicae and Aphis gossypii, among other aphids, have been reported as vectors (Worral et al., 2015). Even though aphid colonization or exuviae were not detected in the flowering and fruit stages, a transient occurrence at early crop stages cannot be rule out. The primary inoculum may originate from diseased plants due to infected seeds, subsequently dispersed by vectors generating foci and aggregates with higher spatial dependance. The severity and incidence levels, contrasted with the phenological events studied, suggest that viral load and infection proportion in seeds is variable among genotypes. Viral transmission may have implications for breeding programs and BCMV resistance studies; therefore, should be investigated.
A. alternata spatial behavior also exhibited contagion progress, although less intense among genotypes, except for Flor de Mayo and Canario genotypes, which, due to higher susceptibility, showed spatial dependence in discontinuous aggregates of 3-8 plants (n= 354.3, σ 2 -p =159.8, σ 2 -s = 514.1). These genotypes had high severity (class 4, 41.5%) at the fruit stage, coinciding with a significant increase and foci coalescence (Figure 4C-B2, B4, 4D- B2 ). Negro Mixteco, Negro Querétaro, and Segregante Oti showed spatial dependence in small random foci of 2-3 plants (n= 103, σ 2 -p = 28.8, σ 2 -s = 87.1), with severity reduction at fruit stage associated with senescence and leaf tissue loss (Figure 4C, 4D- B12 ). Pinto Texcoco, Flor de Mayo, or Garrapato represented genotypes with discontinuous aggregate dispersal pattern of 4-5 plants (n= 58.3, σ 2 -p > 72.9, σ 2 -s = 131.2) and moderate severity (Figure 4C, 4D- B3 ). Vaquita Negro had low severity with uniform dependence (n= 102, σ 2 -p ≈ 0, σ 2 -s = 172.8), however, associated with low covered-plant vigor (0.1) as well as high susceptibility to BCMV (58.1%), therefore, it could be a competition effect rather than tolerance (Figure 4C, 4D- B11 ).
The microclimatic data recorded during the experiment showed minimum relative humidity (RH) between 22 - 51% and RH-maximum >77%; minimum temperature (T) of 11.6 - 15.8 °C and T-maximum of 21.6 - 28.9 °C, and discontinuous post-flowering and fruit rainfall frequency. These conditions are considered optimal for A. alternata mycelial growth and sporulation (Prasad and Ahir, 2013); thus, the dispersal patterns exhibited could represent the real fungus parasitic fitness conditioned by genotype. Consequently, the fungal epidemic potential will be strongly influenced by genetic factors and other contributing abiotic components (Table 4).
Abiotic effects associated with A. alternata infection on bean. Wind ( V ) and hail ( G ) damage simulation was associated with symptoms in 100% of the experimental leaflets, except for Soil ( S ) and the absolute control (Table 4). Necrotic lesions caused by inoculation of A. alternata isolate FPTA2h were reproduced (Figure 2B). Foliar disease severity was directly proportional to conidia concentration applied for each simulated factor. Overall, the G effect induced a higher severity, which ranged in 75.3 - 94.7%, in comparison to V with 66 - 95.7% (Tukey, p = 0.05). Treatments V and G at a 49000 conidia mL-1 concentration (C3) induced 95.3% and 94.7% severity, respectively (Table 4; Figure 2B). Concentrations of 39000 (C2) and 20000 mL-1 (C1) in G induced 80.3% and 75.3% severity, respectively, with not statistical differences (Tukey, p = 0.05). Treatments V -C1 and V -C2 induced a similar severity of 66% and 64.3%, respectively (Table 4; Figure 2B).
Table 4 Percentage of severity in detached bean leaflets of Flor de Mayo genotype (P. vulgaris), inoculated with three conidia concentrations (C) of Alternaria alternata isolate FPTA2h, previously damaged with simulated wind, hail and soil in controlled conditions.
| Tipo de Daño Simulado | Concentración (conidios mL-1) | Porcentaje x de Severidad (± Std. Dev) | |
|---|---|---|---|
| Viento (V) | C1 (20,000) | 66.0 ± 4 d | |
| C2 (39,000) | 64.3 ± 8.5 d | ||
| C3 (49,000) | 95.3 ± 3.5 a | ||
| Granizo (G) | C1 (20,000) | 75.3 ± 4 c | |
| C2 (39,000) | 80.3 ± 4 b | ||
| C3 (49,000) | 94.7 ± 6.1 a | ||
| Suelo (S) | C1 (20,000) | 0.0 (Presencia de biofilm bacteriano) | |
| C2 (39,000) | |||
| C3 (49,000) | |||
| Testigo (T) | Agua destilada Estéril | 0.0 | |
x Treatments with at least one common letter are statistically equal (Tukey, p = 0.05).
A biofilm of colonies, putatively associated with soil-borne pathogenic bacteria was found in S -Ci treatment leaflets (Figure 2S). Despite the bacteria(s) were not identified, the survival of Xanthomonas axonopodis pv. allii and X. axonopodis pv. phaseoli has been reported on bean-fields soil surface, with dispersal capacity due to impact or particle splashing (Torres et al., 2019; Gent et al., 2005). These results confirm that abiotic factors’ effect on infection, contagion, and dispersal of Alternaria and other organisms (Kumar et al., 2021; Lione et al., 2020). Likewise, it suggests that effectiveness of A. alternata inoculum load on beans may depend on secondary factors for infective success, thus could be considered an opportunistic fungus (Armitage et al., 2020). Bean may have evolved efficient structural defense mechanisms during the long plant-pathogen coevolutionary process in America, area of bean domestication (Rendón-Anaya et al., 2017; Bitocchi et al., 2017). Even though a hail event is erratic, the tissue damage by wind in high density crops, such as in P. vulgaris, can be intermittent and determinant for disease (de Langre, 2008).
Conclusions
Appling classical cultural-morphology, genomic, and pathogenic criteria, Alternaria alternata was identified as the causal agent of leaf blight in 12 genotypes of bean (P. vulgaris). Accessions OL229866, OL229867, OL229868, OL229869, and OL229870 were registered in the GenBank. Similarly, through universal primers for Potyvirus and Begomovirus, the association of Potyvirus Bean common mosaic virus with all genotypes was only found. Accessions OL229871, OL229872, OL229873, OL229874, and OL229875 were also registered. In flowering and fruit phenological stages, Negro Perla genotype was the most tolerant with 6.7 - 9.6% and 6.2 - 10% severity associated with BCMV and A. alternata, respectively. Canario was the most susceptible genotype with maximum values of 48.7% and 42.7%. Vigor integration with severity caused by both organisms (IDI) confirmed Negro Perla as the most tolerant variety. Geospatial studies demonstrated differential damage intensity among genotypes and higher BCMV epidemic capacity. A. alternata could be an opportunistic pathogen. A canopy coverage Vigor Index (IV) assisted with single exposure aerial-drone imagery; Integrated Damage Index (IDI) with vigor values and multi-pathogen severity levels; logarithmic-diagrammatic scale at leaf and row-canopy levels; and the App-Monitor® v1.1 Android®, available in PlayStore®, are proposed as novel methodologies to optimize comprehensive breeding programs incorporating phytosanitary factors to complement the phenotypic approach to assess yield components.











 texto en
texto en 


