While the chloroplast genome (plastome) remains highly conserved in most angiosperms (Daniell et al. 2016), there are exceptions characterized by significant structural changes. These variations have been reported in some species-rich groups such as Fabaceae, Passifloraceae, and Malpighiales (Cauz-Santos et al. 2020, Jin et al. 2020, Lee et al. 2021), often observed in parasitic plants (Wolfe et al. 1992, Braukmann et al. 2013, Frailey et al. 2018, Su et al. 2021) or species adapted to extreme environments (McCoy et al. 2008, Silva et al. 2016, Wei et al. 2021). In this scenario, the Cactaceae family serves as a prominent example within the Caryophyllales order. Plastomes in Caryophyllales members usually range from 151 to 155 kb and exhibit the typical quadripartite structure divided into a large single copy (LSC), a small single copy (SSC), and two inverted repeats (IRa and IRb) regions (Yao et al. 2019). Nevertheless, numerous structural arrangements have been identified within Cactaceae. These arrangements involve plastome reduction derived from expansion/reduction or loss of the inverted repeat regions (IRs), and gene losses (Sanderson et al. 2015, Majure et al. 2019, Köhler et al. 2020, Morais Da Silva et al. 2021, Köhler et al. 2023, Yu et al. 2023).
Chloroplast genomes within Cactaceae exhibit a wide range in length, spanning from 107 to 162 kb, and their IRs vary from around 350 bp to 37 kb (Solórzano et al. 2019, Köhler et al. 2023, Yu et al. 2023). It is worth noting that Carnegiea gigantea (Engelm.) Britton & Rose (the saguaro) and Lophocereus schoottii (Engelm.) Britton & Rose, lack the IRs (Sanderson et al. 2015, Solórzano et al. 2019), as well as other Opuntioideae such as Quiabentia verticillata (Vaupel) Borg (Köhler et al. 2020). Within the genus Mammillaria Haw., nine chloroplast genomes have been sequenced with lengths ranging from 107 to 116 kb and three distinct structural variations (Solórzano et al. 2019, Hinojosa-Alvarez et al. 2020, Yu et al. 2023).
In Mammillaria, the structure of the chloroplast genome displays variation. Solórzano et al. (2019) assembled chloroplast genomes for seven species representing three out of the eight subgenera proposed by Hunt et al. (2006): Krainzia, Mammillaria, and Phellosperma. Interestingly, species from different subgenera share the same chloroplast genome structure (Solórzano et al. 2019). Consequently, the different chloroplast genome structures seem to not be related to the morphology-based classification. Given the variation of the plastome structure in Mammillaria, other unknown structures may exist in the remaining subgenera or in other groups within the subgenus Mammillaria.
Plastome sequences and information are lacking for the Mammillaria series Stylothelae, which is part of M. subg. Mammillaria and encompasses 19 taxa (Hunt 2016, González-Zamora et al. 2023, Ortiz-Brunel et al. 2023). This series can be distinguished by the presence of axillary bristles, slightly embedded flowers and fruits, generally uncinated central spines, and the production of Luethy’s alkaloid. The combination of these characters differentiates Stylothelae from the other series (Fitz-Maurice & Fitz-Maurice 2006, Hunt et al. 2006). Additionally, all species of M. series Stylothelae share the absence of the rpl16 intron, which is proposed as a synapomorphy of the group (Butterworth & Wallace 2004, Butterworth et al. 2007). This could be an indicator of chloroplast structure variation. Based on this evidence, we anticipate that species of M. subg. Mammillaria series Stylothelae may exhibit a different chloroplast genome structure.
Analyses of chloroplast genome structures and phylogenetic hypothesis based on whole chloroplast sequences have demonstrated their utility in elucidating phylogenetic relationships within Mammillaria and its related genera (Solórzano et al. 2019, Breslin et al. 2021, Chincoya et al. 2023). Our objective was to sequence and assemble the plastome of M. bocasana Poselg. and M. erythrosperma Boed., which belong to M. subg. Mammillaria series Stylothelae (Hunt et al. 2006). Additionally, we also compared their structures with those previously described for the genus to gain insights into a broad overview of plastome evolution within the genus.
Material and methods
Sampling. Two living individuals of Mammillaria bocasana (J. P. Ortiz-Brunel 922, IBUG) and two of M. erythrosperma (J. P. Ortiz-Brunel 410, IBUG) (herbarium acronym according to Thiers 2023) were collected nearby their type locality (Figure 1). The morphology of the specimens was compared with the morphological descriptions to corroborate their identity (Bravo-Hollis & Sánchez-Mejorada 1991, Reppenhagen 1991, Fitz-Maurice & Fitz-Maurice 2006). We preserved the plants in a greenhouse at the University of Guadalajara until tissue collection.
DNA extraction and sequencing. Using freshly collected tissue from the tubercles of only one individual per species, chloroplasts were isolated and cpDNA extracted as described in Shi et al. (2012), with slight modifications. The cpDNA quantity and quality were evaluated with a Qubit 3.0 and NanoDrop 2000 (Thermo Fisher Scientific, Waltham, Massachusetts), respectively. We prepared the libraries using the Ion Plus Fragment Library Kit (Thermo Fisher Scientific) and selected DNA fragments of approximately 250 bp using an E-Gel Sizeselect Agarose Gel (Thermo Fisher Scientific). Sequencing of single-end reads of 250 bp was performed on a Personal Genome Machine (Thermo Fisher Scientific) in the Laboratorio Nacional de Identificación y Caracterización Vegetal (LaniVeg) at the University of Guadalajara.
Chloroplast genome assembly. Raw reads quality was assessed in the FastQC v. 0.11.7 program (Andrews 2010). Then, we used the Trimmomatic tool (Bolger et al. 2014) to discard reads with low quality (PHRED quality score < 15) using the leading, trailing and avgqual tools. The resulting reads were de novo assembled into contigs following the Fast-Plast pipeline (McKain 2017) with slight modifications. Reads were mapped against the genome of Mammillaria pectinifera F.A.C. Weber as a reference using Bowtie2 v. 2.5.1 (Langmead & Salzberg 2012) under the very-sensitive-local parameter set to filter for chloroplast-like sequences. These filtered reads were de novo assembled using SPAdes 3.15.0 (Bankevich et al. 2012) using k-mer sizes of 21, 35, 57, and 89 with the “only-assembler” option. The assembled contigs were merged and extended using the “afin” script available from the Fast-Plast program using default parameters with 50 extension loops and the chloroplast-mapped reads. Once a single contig was reached, the sequence_based_ir.pl script packaged with Fast-Plast was used to find putative IR regions. We used Sequencher v. 4.1.4 (Gene Codes) to verify the IRs. A final coverage analysis to verify the accuracy of our assemblies was conducted using scripts from Fast-Plast and supported by Jellyfish 2 (Marçais & Kingsford 2011). Gene annotation was performed in the GeSeq platform (Tillich et al. 2017) and every annotation was manually confirmed. The chloroplast genome circular representation was produced with OGDraw v. 1.3.1 (Greiner et al. 2019).
Chloroplast genome structure comparison. We selected one sequence for each known chloroplast genome structure in Mammillaria (Solórzano et al. 2019) for comparison to our new assemblies. Mammillaria pectinifera represented the structure 1 (S1), M. crucigera Mart. characterized the structure 2 (S2), and M. zephyranthoides Scheidw. corresponded to the structure 3 (S3) (Solórzano et al. 2019). These representative chloroplast genomes were converted into linear genomes and then aligned to our M. bocasana and M. erythrosperma plastomes. All genomes were aligned using the plastome of M. bocasana as the reference in MAUVE v. 2.4.0 (Darling et al. 2004) with the Progressive Mauve Tool using the default parameters. GenBank accession numbers are indicated in Table 1.
Table 1 List of the species used in the analyses.
| Species | GenBank accession number | Infrageneric classification sensu Hunt et al. (2006) |
|---|---|---|
| Carnegiea gigantea | NC_027618.1 | |
| Mammillaria albiflora | MN517610.1 | Subgenus Krainzia, series Herrerae-Pectiniferae |
| M. bocasana | OR863748 | Subgenus Mammillaria, series Stylothelae |
| M. crucigera | MN517613.1 | Subgenus Mammillaria, series Supertextae |
| M. erythrosperma | OR863749 | Subgenus Mammillaria, series Stylothelae |
| M. huitzilopochtli | MN517612.1 | Subgenus Mammillaria, series Supertextae |
| M. pectinifera | MN519716.1 | Subgenus Krainzia, series Herrerae-Pectiniferae |
| M. solisioides | MN518341.1 | Subgenus Krainzia, series Herrerae-Pectiniferae |
| M. supertexta | MN508963.1 | Subgenus Mammillaria, series Supertextae |
| M. zephyranthoides | MN517611.1 | Subgenus Phellosperma |
Phylogenetic analysis. We downloaded seven chloroplast genome sequences of Mammillaria from GenBank published by Solórzano et al. (2019). These sequences represented the three different chloroplast genome structures within the genus. The plastome of Carnegiea gigantea (Sanderson et al. 2015) was included as an outgroup. Detailed GenBank accession numbers for all sequences used in the analysis are listed in Table 1. We aligned the ten complete plastome sequences using MAFFT v. 7.52 (Katoh et al. 2019) with default parameters. A maximum likelihood (ML) search was executed in MEGA v. 11 (Tamura et al. 2021), employing the GTR + G + I model. Supporting branch values were obtained through 1,000 bootstrap replications. To discard a possible influence of the chloroplast structures in the results, we performed two additional phylogenetic analyses with the same parameters but different datasets. The first analysis used a matrix in which the 21 kb inversion block was reverted for M. bocasana and M. erythrosperma. In the second one, we filtered for 54 CDS regions shared among all taxa (Table S1). All phylogenetic analyses were performed with the same parameters.
Results
Chloroplast genome structure comparison. The chloroplast genomes of Mammillaria bocasana and M. erythrosperma exhibited a quadripartite structure, including an LSC, an SSC, and two small IRs. The chloroplast genome of M. bocasana was 107,368 bp long with an LSC of 75,290 bp, an SSC of 28,896 bp, and two IRs of 1,591 bp. Mammillaria erythrosperma chloroplast genome was 108,069 bp long, from which 76,393 bp conformed the LSC, the SSC was of 28,402 bp, and two IRs of 1,637 bp (Figure 2). The Guanine-Cytosine (GC) content was 37 % in M. bocasana and 36.6 % in M. erythrosperma.
Both plastomes shared identical gene content and order with 108 protein coding genes, tRNAs, and rRNAs (Figure 2, Table 2). Ten of those genes were pseudogenized in Mammillaria bocasana and 11 in M. erythrosperma. The latter exhibited pseudogenization of the rps16 gene, resulting from a partial loss of the first exon. The IRs of both species contained rpl2, pseudogene rpl23, and trnI-CAU with IR lengths of 1,591 bp in M. bocasana and 1,637 in M. erythrosperma. In both plastomes, the IRa was delimited by the rps19 and the ycf2 genes and the IRb by the ndhB (pseudogene) and trnH-GUG genes. Both plastomes lacked functional NADH dehydrogenase-like (NDH) complex (ndh) genes, though we detected only pseudogenes of ndhB, ndhD, and ndhF.
Table 2 Gene content of the chloroplast genome of M. bocasana and M. erythrosperma. Ψ indicates a pseudogene in both species; *Complete gene in M. bocasana, pseudogene in M. erythrosperma; **One copy functional and the other a pseudogene in both species.
| Genes group | Name | Number | |
|---|---|---|---|
| Photosynthesis | Photosystem I | psaA, psaB, psaC, psaI, psaJ | 5 |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 | |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | 6 | |
| NADH dehydrogenase | ndhBΨ, ndhDΨ, ndhFΨ, | 3 | |
| Cytochrome complex | petA, petB, petD, petG, petL, petN | 6 | |
| Rubisco Large Subunit | rbcL | 1 | |
| Acetyl-CoA carboxylase beta subunit | accDΨ | 1 | |
| Genetic expression control | Ribosomal large subunit proteins | rpl2 (2), rpl14, rpl16, rpl20, rpl22, rpl23 (2)Ψ, rpl32, rpl33Ψ, rpl36 | 11 |
| RNA polymerase subunits | rpoA, rpoB, rpoC1, rpoC2 | 4 | |
| Ribosomal small subunit proteins | rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps15, rps16*, rps18, rps19 | 12 | |
| Ribosomal RNA | rrn16, rrn23, rrn4.5, rrn5 | 4 | |
| Transfer RNA | trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU (2), trnI-GAU, trnK-UUU, trnL-CAA, trnL-UAA, trnL-UAG, trnM-CAU, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnW-CCA, trnY-GUA | 29 | |
| Other | Conserved Open Reading Frames (ORF) | ycf1, ycf2, ycf3, ycf4Ψ, ycf68Ψ | 5 |
| Cytochrome C synthesis | ccsA | 1 | |
| Chloroplast envelope membrane protein | cemA | 1 | |
| Maturase | clpP (2**), matK | 3 | |
| Translation initiation factor | infA | 1 | |
| Total | 108 |
When compared to other Mammillaria plastome structures, the progressive MAUVE alignment identified a new chloroplast genome structure (S4) characterized by a unique inversion within the LSC region in both species of Mammillaria series Stylothelae (Figure 3). This inversion was 21,248 bp long in M. bocasana and 21,469 bp in M. erythrosperma and encompassed 21 coding regions, extending from petG to atpE genes (Figure 2). The majority of these inverted genes are associated with photosynthetic functions (Table 2). The analyzed species, that correspond to the S1, S2, and S4, had all the other gene blocks in the same order and arrangement. However, the alignment showed remarkable reductions and inversions in the S3, compared with all the other structures.
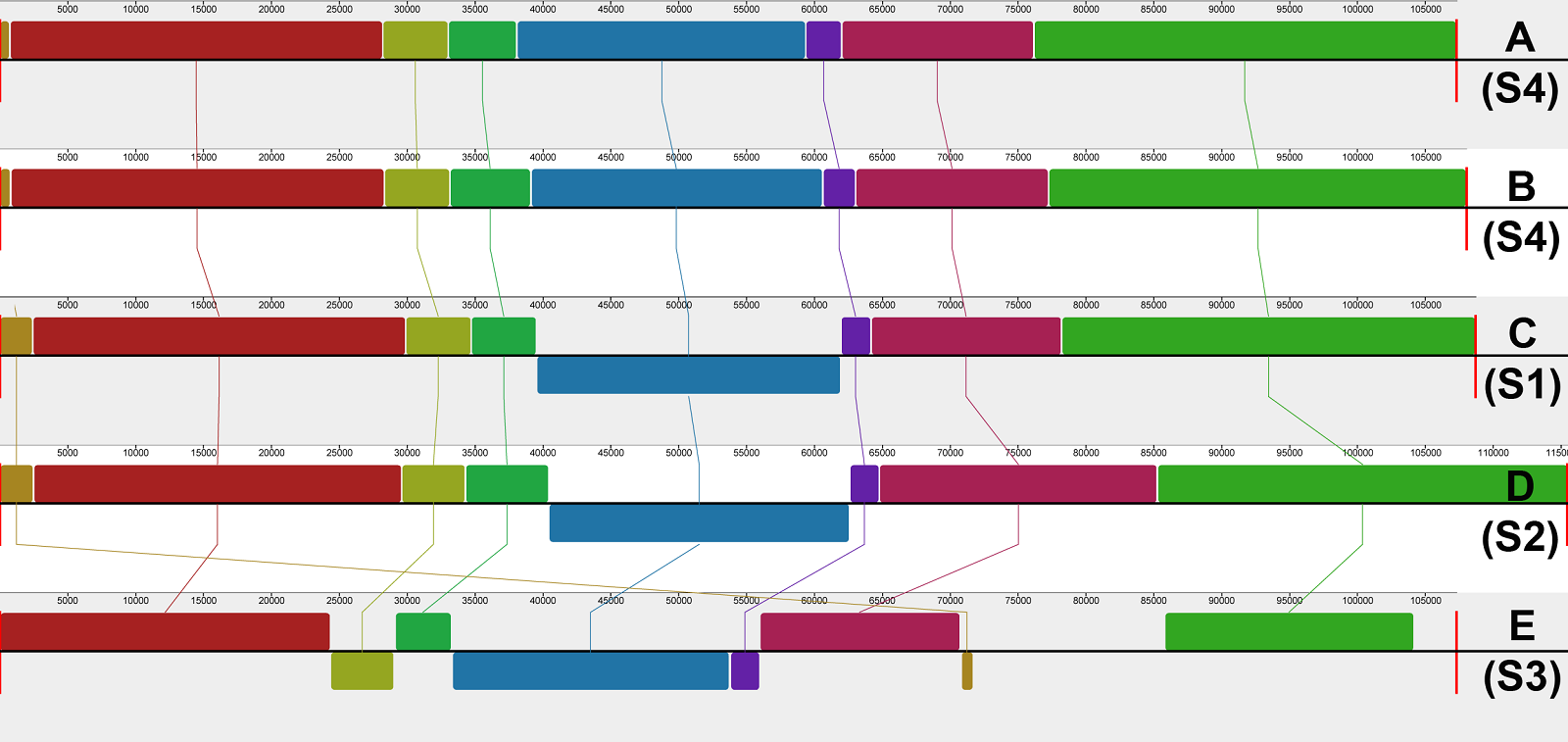
Figure 3 MAUVE alignment. A) Mammillaria bocasana, B) M. erythrosperma, C) M. pectinifera, D) M. crucigera, E) M. zephyranthoides. The codes S1, S2, and S3 represent the chloroplast genome structures found by Solórzano et al. (2019) and S4 indicates the new structure found in this work.
Phylogenetic analysis. The chloroplast genome recovered robust phylogenetic relationships, with bootstrap support values (BS) exceeding 95 % for all tree branches (Figure 4). The analyses with the complete plastomes, the complete plastomes with the 21 kb reverted block, and the shared CDS, recovered mostly the same topology. These differed in the placement of Mammillaria albiflora and M. zephyranthoides, and the relationships within M. series Supertextae (Figures 4, S1, S2). Here, we used the phylogeny inferred with the complete plastomes to describe and discuss our results (Figure 4). The resulting phylogeny placed our two plastome sequences in a monophyletic group within all other Mammillaria species (BS = 100 %). Interestingly, species with the same chloroplast structure were dispersed across different clades (Figure 4), except for Mammillaria bocasana and M. erythrosperma, which had a sister species relationship with strong support (BS = 100 %). Further, this clade was sister to a group that included species from M. subg. Mammillaria series Supertextae (M. supertexta, M. crucigera and M. huitzilopochtli) and M. subg. Krainzia (M. pectinifera and M. solisioides). The phylogeny did not support the monophyly of Mammillaria subg. Mammillaria nor M. subg. Krainzia (Figure 4, Table 1). While M. subg. Mammillaria appeared to be paraphyletic, M. series Supertextae (BS = 100 %) and M. series Stylothelae (BS = 100 %) formed two monophyletic groups.
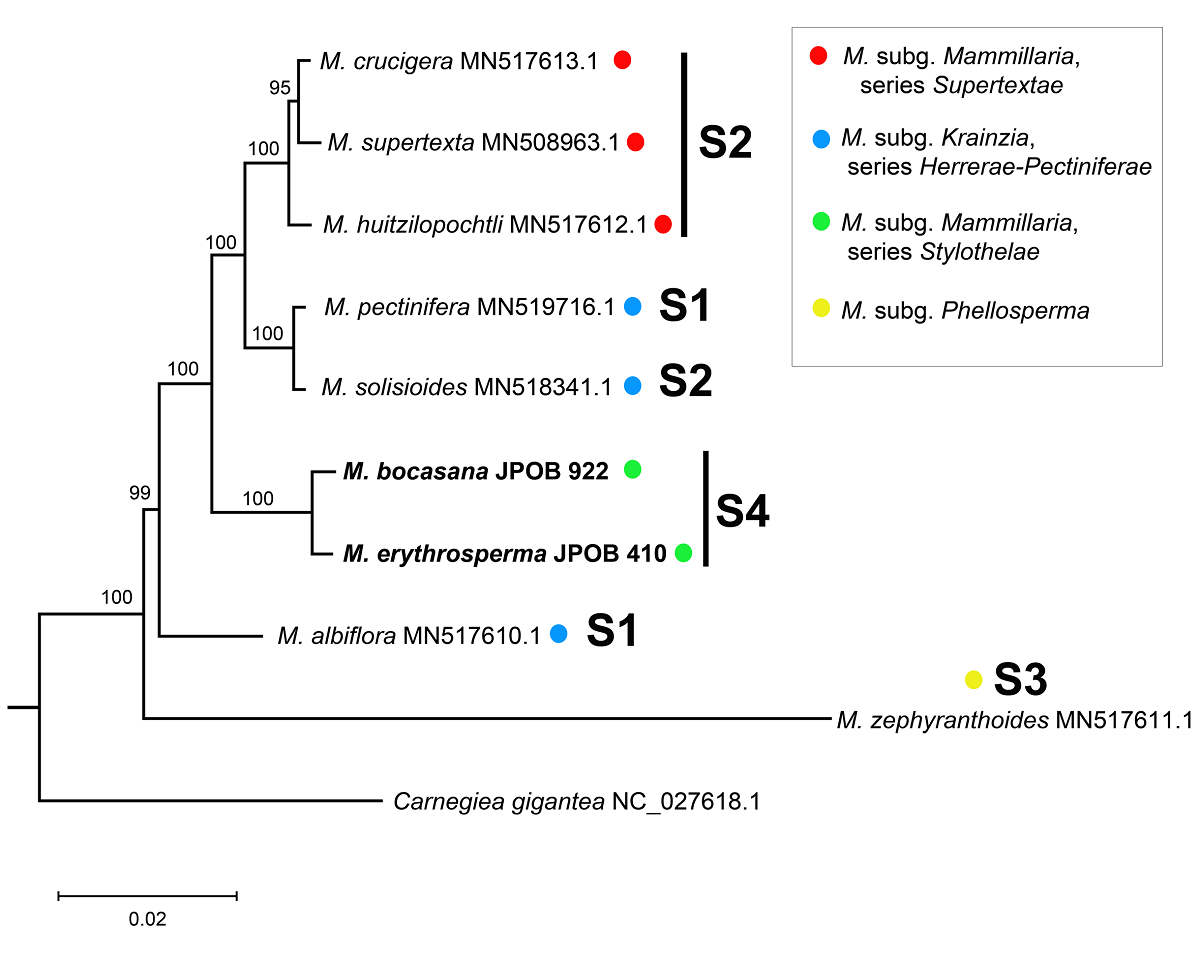
Figure 4 Phylogenetic analysis based on Maximum Likelihood. The numbers above the branches represent the Bootstrap support values. Codes for the structures (S) are the same as in Figure 3.
Discussion
Chloroplast genome structure comparison. The chloroplast genome sequences of Mammillaria bocasana and M. erythrosperma (Mammillara series Stylothelae) exhibit an undescribed plastome structure (S4) (Figure 2). This new structure shares similar gene content and arrangement with the S1, represented by M. pectinifera (Solórzano et al. 2019). It differs, however, by a ~ 21 kb inversion within the LSC that had not been reported in other Mammillaria species (Figure 3). The inversion contains mainly protein coding genes related to photosynthesis. On the other hand, the IRs of M. bocasana and M. erythrosperma (S4) were similar in length and gene content to those of M. albiflora and M. pectinifera (S1) (Solórzano et al. 2019). The distinctive features of this new structure are the inversion within the LSC and the presence of the rpl2 gene in the IRs instead of a partial sequence of the ycf2 gene (Table 3). Our results support the idea that trnI-CAU is involved in reconfiguring of IRs in Mammillaria (Solórzano et al. 2019). Boundary shifts seem to result from gene rearrangements, but further research is needed.
Table 3 Length and gene content of the inverted repeat regions of Mammillaria. The symbol Ψ indicates a pseudogene. The S1 to S3 structures are those reported by Solórzano et al. (2019). S4 denominates the structure found in this work.
| Structure | Species | IR length (bp) | IR genes content |
|---|---|---|---|
| S1 | Mammillaria albiflora, M. pectinifera | 1,348; 1,544 | rpl23, trnI-CAU, ycf2 |
| S2 | Mammillaria crucigera, M. huitzilopochtli, M. solisioides, M. supertexta | 14,522; 14,488; 14,428; 14,490 | trnQ-UUG, rps16, trnK-UUU, matK, psbA, trnH-GUG, rpl2, rpl23, trnI-CAU, ycf2 |
| S3 | Mammillaria zephyranthoides | 28,252 | psbA, trnH-GUG, trnI-CAU, ycf2, ndhB, rps7, rps12, trnV-GAC, rrn16, trnI-GAU, rrn23, rrn4.5, rrn5, trnR-ACG, trnN-GUU, ndhF, rpl32 |
| S4 | Mammillaria bocasana, M. erythrosperma | 1,591; 1,637 | rpl2, rpl23Ψ, trnI-CAU |
Compared with other Cactaceae, the Mammillaria plastomes display high variation associated with gene translocations and inversions (Solórzano et al. 2019, Yu et al. 2023). Plastome variation is common in Cactaceae and typically involves changes within the IRs. This variation is given by gene rearrangements, expansion and contraction of coding and non-coding regions, and changes in the boundaries of the IRs (Yu et al. 2023). Palmer (1986) and Walker et al. (2015) suggested that the IR could induce isomers, particularly within the SSC. This phenomenon is common in Cupressaceae (Guo et al. 2014, Qu et al. 2017) and was recently discovered within the LSC in Cactaceae (Yu et al. 2023). In cacti, the frequence of isomers is estimated around 1 % (Yu et al. 2023). Another source of plastome variation is the high presence of short repeat sequences that promote chloroplast structure differentiation mediated by intramolecular recombination (Ruhlman et al. 2017, Qu et al. 2017). When comparing the plastomes in the MAUVE alignment, we found short sequence repeats near the boundaries of the 21 kb inverted block of genes within the LSC of M. bocasana and M. erythrosperma. High short repetitive regions are common within the LSC of Mammillaria plastomes (Chincoya et al. 2020). It is possible that repetitive sequences in Mammillaria may serve as recombination points and cause the rearrangements. As stated and demonstrated by Yu et al. (2023) more studies are needed to confirm this or to discard the presence of plastome isomers. Given this, it is possible that some Mammillaria plastome structures are merely isomers, but further evaluation is needed.
In Cactaceae, the length of chloroplast genomes exhibits high variability due to gene losses, gene duplications, pseudogenization, and expansions/contractions of the IRs (Solórzano et al. 2019, Köhler et al. 2023, Yu et. al. 2023). Mammillaria is known for having some of the shortest plastomes within the family, and our findings were consistent with this pattern. In the plastome of M. bocasana and M. erythrosperma, we observed the pseudogenization of the following genes: accD, ndhB, ndhD, ndhF, rpl23 (both copies), rpl33, rps16, ycf4, ycf68, and clpP (one copy) (Table 2). All these pseudogenes have been previously found in other species of the genus (Solórzano et al. 2019) and are common within Cactaceae (Yu et al. 2023). The plastomes of M. bocasana (S4) and M. zephyranthoides (S3) are the shortest among Mammillaria (~ 107 kb), but they differ from each other in their gene content (108 and 130, respectively) and arrangement, suggesting a different evolutionary pathway. According to Chincoya et al. (2023), the divergence of most Mammillaria clades occurred ~ 4.5 Mya, which implies a recent diversification of chloroplast structures within the genus. It is necessary to assemble more Mammillaria plastomes to trace an accurate evolutionary history.
The evolutionary implications of losing ndh genes are not fully understood. These genes play a role in the cyclic electron flow of ATP production (Martín et al. 2009, Strand et al. 2019). The partial or complete loss of the ndh genes suite is common in gymnosperms (Braukmann et al. 2009, Martín & Sabater 2010) and frequently occurs in angiosperms (Blazier et al. 2011, Sun et al. 2017, Sun et al. 2018, Könyves et al. 2021, Mower et al. 2021, Cao et al. 2022). In Cactaceae, the complete loss or pseudogenization of multiple ndh genes is common (Sanderson et al. 2015, Solórzano et al. 2019, Morais da Silva et al. 2021, Köhler et al. 2023, Yu et al. 2023). In general, it is not yet clear whether these genes have been transferred to the mitochondrial or nuclear genomes or if there are alternative metabolic pathways that compensate for their absence (Lin et al. 2015, Ruhlman et al. 2015, Sanderson et al. 2015, Ranade et al. 2016, Strand et al. 2019). These genes appear to be dispensable under favorable conditions but become crucial when plants are exposed to abiotic stress conditions (Ruhlman et al. 2015, Lin et al. 2017, Sabater 2021). All Mammillaria species lacking the ndh genes inhabit arid or semiarid regions and it remains unknown how they grow in harsh environments and compensate for the absence of some or all of these genes.
The de novo assembled plastomes of Mammillaria bocasana and M. erythrosperma lacked the rpl16 intron. However, the gene seems to be completely functional because only the main intron is excised and a complete gene remains (Butterworth et al. 2007). The same case has been rarely documented in Amaryllidaceae, Geraniaceae, Goodeniaceae, Papaveraceae, and Plumbaginaceae, but it is infrequent even within them (Campagna & Downie 1998, Zhang et al. 2020, Kim et al. 2023). Consequently, this feature is considered a robust signal of common ancestry (Campagna & Downie 1998). Up to this point, all the other chloroplast genomes known for Mammillaria have the rpl16 intron. Therefore, it is highly plausible that the absence of the rpl16 intron could be a synapomorphy for M. series Stylothelae, as suggested by Butterworth et al. (2007). To evaluate this, additional taxa sampling is necessary, including recently described species within the series (González-Zamora et al. 2022, 2023, Ortiz-Brunel et al. 2023).
Phylogenetic analysis. The ML tree confirmed the inclusion of our newly sequenced chloroplast genomes within Mammillaria (Figure 4). Mammillaria subg. Mammillaria is paraphyletic, partly due to the inclusion of M. pectinifera and M. solisioides, which belong to M. subg. Krainzia. Similar results have been reported in recent, more comprehensive studies, indicating the need for further research (Chincoya et al. 2023). Our results agreed with the monophyly of series Supertextae, which was also identified by Cervantes et al. (2021). In all three phylogenetic analyses performed with different datasets, Mammillaria bocasana and M. erythrosperma grouped as sister to the clade containing the subgenera Mammillaria and Krainzia. It is possible that M. series Stylothelae is monophyletic, as well as other series within M. subg. Mammillaria. Taxon sampling was limited in the present and previous works, and thereby only limited conclusions can be drawn until denser sampling can be done.
Our phylogenetic analyses support that using complete chloroplast genome sequences or only the shared CDS can produce well-supported hypotheses. An independent study based solely on chloroplast protein-coding genes yielded a similar topology (Solórzano et al. 2019). In this study, we performed a phylogenetic analysis with the original full plastomes aligned, other with the ~ 21 kb inversion of M. bocasana and M. erythrosperma reverted, and another only with 54 shared CDS for all taxa. The full original plastomes dataset produced a better resolved phylogeny, but the hypothesis generated with the inverted block reverted retrieved the same topology and support (Figures 4, S1). However, using the CDS dataset, the placement of M. albiflora and M. zephyranthoides was different (Figures 4, S2). Any of the three approaches is useful in establishing insights into the evolutionary history of the Mammillaria chloroplast genomes. However, it is necessary to increase the taxon sampling to test if some structures have a unique origin (Figure 4). Different chloroplast genome structures have been identified in some species-rich groups with significant morphological variation (Cauz-Santos et al. 2020, Köhler et al. 2020, Lee et al. 2021). With the discovery of a fourth structure characterizing until now the Mammillaria series Stylothelae, it becomes evident that more extensive taxon sampling across Mammillaria is required. Other chloroplast structures might exist within the genus.
Supplementary material
Supplemental data for this article can be accessed here: https://doi.org/10.17129/botsci.3446.











 nueva página del texto (beta)
nueva página del texto (beta)





