Introduction
The microbial diversity present in ecosystems is a very important component and a determining factor of biological equilibrium (Ofek-Lalzar et al., 2014). The alteration undergone in an ecosystem has an impact on different scales and on physicochemical and biological parameters (Lareen, Burton, & Schäfer, 2016; Ofek-Lalzar et al., 2014). The characterization of microorganisms and their relative abundance in different environments provides very important information on both the preservation of this ecosystem and the potential use of native microorganisms (Panke-Buisse, Poole, Goodrich, Ley, & Kao-Kniffin, 2015).
In Mexico, climatic, orographic and water factors result in a wealth of environments. There are reports of isolation of different types of microorganisms from soils of various ecosystems in the country (Rodríguez-Zaragoza et al., 2008; Vásquez-Murrieta, Govaerts, & Dendooven, 2007). The main limitations for carrying out these processes have been the difficult access to the places to obtain samples and the multiple requirements for the isolation and identification of the microorganisms. Only 1 % of the microorganisms in a soil sample are cultivable in laboratory media (Ofek-Lalzar et al., 2014; Panke-Buisse, Lee, & Kao-Kniffin, 2017; Vaz-Moreira, Nunes, & Manaia, 2014); therefore, the application of a technique that does not require isolation and culture would allow the characterization of a larger fraction of the soil microbiology.
Soil should be conceived as a living entity, in which each of the microorganism groups function as organs of a biological entity, with specialized functions, but at the same time interconnected (Lareen et al., 2016; Nurulita, Adetutu, Gunawan, Zul, & Ball, 2016; Poosakkannu, Nissinen, & Kytöviita, 2017). Being a living organism, the soil can become diseased, and diagnosing the condition requires a series of specific analyses (Nurulita et al., 2016; Rota et al., 2013). The traditional approach is to characterize the physicochemical parameters of the soil to detect its deficiencies; however, the new paradigm is to evaluate it microbiologically and establish the degree of alteration of the processes that must occur naturally (Lareen et al., 2016; Panke-Buisse et al., 2017).
The impact of polluting substances on the environment has been studied for a long time; the most significant is that which occurs from fossil fuels. Since the Exxon Valdez oil spill in 1989, studies have been carried out on the ability of microorganisms to degrade this type of pollutant. The results have been favorable, thanks to the ability of microorganisms to adapt to the most severe conditions possible (Atlas & Bragg, 2009; Bacosa, Suto, & Inoue, 2011; Feng et al., 2007; Fukuyama, Shigenaka, & Coats, 2014; Rodriguez-R et al., 2015; Sharifi, Van Aken, & Boufadel, 2011). Because many fuel spills occur in soil and water, the option is to use microorganisms that adapt to these two conditions and that can pass from one medium to another while conserving their degrading ability, an intrinsic characteristic that only microorganisms are capable of developing (Boopathy, Shields, & Nunna, 2012; Bouchez-Naïtali & Vandecasteele, 2008; Guo et al., 2012; Patel, Cheturvedula, & Madamwar, 2012; Stepanyan & Voskoboinikov, 2006).
This study analyzed the ability of groups of microorganisms with stable symbiotic associations, obtained from a cloud forest ecosystem in Mexico, to degrade aromatic compounds (benzene, toluene, ethylbenzene and anthracene), the main pollutants present in fossil fuels. This type of forest is characterized by a mixed composition of temperate climate species in the canopy, tropical and subtropical ones in the sub-canopy and understory, and an abundance of mosses, epiphytes and tree ferns that confer ample biodiversity to this ecosystem; in addition, it is geologically associated with the Paleozoic Era. For this reason, the territory has a rich soil biology, which can be promising for the degradation of aromatic hydrocarbons.
Materials and methods
Vegetation sampling was carried out in the Sierra Madre del Sur in the state of Oaxaca, Pochutla district, at an elevation of 1 300 m, where there are archipelagos of the mesophilic mountain forest (Gual-Díaz & Rendón-Correa, 2014). After recording the number of trees, species abundance was calculated considering 10 sites located in different plots. To corroborate the data obtained, Margalef’s index was used for specific richness and Simpson’s for abundance. Margalef’s index is based on the relationship between the number of species (S) and the total number of individuals observed (N) and is defined as:
where, p i is the proportional abundance of species i; that is, the number of individuals of species i divided by the total number of individuals in the sample.
Obtaining groups of microorganisms
In the cloud forest archipelagos, 50 soil microbiome samples were collected in different areas of the ecosystem, as shown in Table 1. The methodology consisted of collecting soil, roots and leaves close to the trees, whose phenotypic characteristics are resistance and prosperity in inhospitable environments. Simple random sampling was carried out in sites with undisturbed vegetation, where the diversity of plants at the site and the amount of leaf litter on the ground were considered, starting from the hypothesis that the specimens of emerging trees should correspond to a biodiversity pattern in the soil.
The samples were cultured in a medium with a mixture of polysaccharides, in the facilities of the organic fertilizer and vermiculture production module at Autonomous Chapingo University’s San Ignacio Field. After the development of the microbiota in the culture systems, samples were taken for genetic identification.
Table 1 Location of soil microbiome samples in a cloud forest of the Sierra Madre del Sur in the state of Oaxaca, Mexico.
| Extraction region | Microorganism groups | Latitude | Longitude |
|---|---|---|---|
| Region I | F1-F10 | 16° 03´ 27.5´´ N | 96° 37´ 08.7´´ W |
| Region II | F11-F20 | 15° 59´ 06.2´´ N | 96° 13´ 57.7´´ W |
| Region III | F21-F30 | 15° 59´ 11.3´´ N | 96° 42´ 2.8´´ W |
| Region IV | F31-F40 | 16° 09´ 56.8´´ N | 97° 29´ 45.1´´ W |
| Region V | F41-F50 | 16° 01´ 00.2´´ N | 96° 34´ 49.5´´ W |
DNA Extraction and sequencing of amplicon libraries
Genetic material was extracted in accordance with the protocol indicated by the manufacturer of PowerSoil® (MO BIO Laboratories, Inc). The DNA was amplified by PCR (Polymerase Chain Reaction), using 50 μL of 1xPCR buffer (5mM KCl, 1mM Tris-HCl, pH 8.0), 2 mM MgCl2, 0.2 mM of each primer 16S rDNA with 1356 bp product), 0.2 mM dNTP, 0.05 U of recombinant Taq polymerase (Thermo scientific EP0402) and 10 ng of total nucleic acids. The PCR program was 25 cycles (30 s, 95 °C; 30 s, annealing temperature for primers; 30 s, 72 °C] in a GeneAmp PCR System 2700 thermocycler (Applied Biosystems, USA).
The amplified material was characterized by massive ion semiconductor sequencing (Ion Torrent equipment, USA) and the results were processed through a bioinformatic analysis to obtain the sequences and make the corresponding identification (García-Mena et al., 2016).
Packed bioreactor test
A continuous-flow borosilicate bioreactor was built, measuring 10 cm in diameter and 70 cm in length, in an isothermal configuration. The fixed bed was made from sterile, additive-free, pharmaceutical-grade cotton fiber. The continuous flow was maintained with a diaphragm dosing pump (Seko brand, USA), from 0 to 100 % flow, with capacity to allow a volumetric flow of 500 mL·min-1. The bioreactor feed culture was prepared in a 20-L glass container, sterile and with continuous aeration by means of a pump (ELITE brand, USA) with a capacity of 60 L·h-1 of air. The receptacle contained 20 L of minimum medium (1.2 mL·L-1 FeCl2 0.1 %, KH2PO4 0.5 g· L-1, MgCl2 0.4 g·L-1, NaCl 0.4 g·L-1, NH4Cl 0.4 g·L-1, CaCl2 0.05 g·L-1, 1mL·L-1 trace element solution [ZnSO4 10 mg·L-1, MnCl2 3.0 mg·L-1, H3BO4 30 mg·L-1, CoCl2 20 mg·L-1, CuCl2 1.0 mg·L-1, NiCl2 2.0 mg·L-1, Na2MoO4 3.0 mg·L-1]; all analytical-grade reagents were Fermont brand) enriched with glucose (2 g·L-1). This medium was inoculated with 2 mL of each of the 50 groups of microorganisms and developed up to the exponential growth phase, for five days at 30 °C. The microorganism count during the test in the feeding system was made with 3M™ brand Petrifilm™ plates for aerobes, according to the methodology indicated by the manufacturer. The pumping system was connected to the bioreactor with a flow of 50 mL·min-1, allowing the establishment and formation of a microbiological film at a temperature of 30 °C.
Once the biofilm was formed, the flow of the glucose-enriched medium was interrupted and the problem solution was fed, consisting of a mixture of the pollutants benzene, toluene, ethylbenzene and anthracene, with a concentration of 1 000 mg·L-1 each. The solution was fed to the bioreactor, taking samples every 5 h, to analyze the concentration of the pollutants present by HPLC-PDA. At the end of the experiment, the bioreactor packing was extracted and divided into equal parts with a length of 10 cm in order to study the behavior of the microorganisms along the system by means of characterization by genetic sequencing.
The mineralization achieved during the process was measured through the analysis of total organic carbon (TOC), inorganic carbon (IC) and total carbon (TC) in General Electric brand equipment (Innova model, USA) with a self-sampling device. The measurement conditions were 5 % sodium persulphate as oxidant and 15 % phosphoric acid. The pollutant mixture used during the test was determined in a HPLC system (Perkin Elmer brand, Flexar model, USA) with a Phenyl column 25 cm long by 4.6 mm in diameter, with particle size of 5 μm, in reverse phase, coupled to a PDA (photodiode array) detector. The working conditions were 95:5 acetonitrile:water and 1.0 mL·min-1 flow in isocratic regime. The preparation of the sample for injection into the chromatograph required a SPE (solid phase extraction) system, using 2-mL C18 cartridges, with a speed of 23 to 26 drops per minute. The cartridge was conditioned with 5 mL of HPLC-grade methanol, followed by loading of the sample, whose required volume ranged from 1 to 100 mL; subsequently, the cartridge was washed with 5 mL of distilled water, and elution of the components was carried out with 5 mL of HPLC-grade acetonitrile.
Results and discussion
Ninety-nine tree species used as shade trees in coffee plantations were counted, many of which are endemic; a total of 1 140 individuals were recorded. Predominant species included pines (Pinus spp.), oaks (Quercus spp.), sweetgum (Liquidambar spp.), magnolias (Magnolia spp.), caudillo (Oreomunnea mexicana [Standl.] J.-F. Leroy), Mexican hand tree (Chiranthodendron pentadactylon Larreat.) and tree ferns (Cyathea spp.). Margalef's diversity index was 13.92 and Simpson's was 0.96. The former indicates that the archipelagos of the mountain cloud forest in the Loxicha region are composed of a wide diversity of variable composition in number of species, and the fact the Simpson’s index is close to 1 ratifies that the site is diverse; the results reveal high biodiversity and abundance of each plant species. This was the criterion used in the selection of areas for microbial sampling and genetic sequencing of soil bacteria.
The sequencing results are shown in Figure 1. In all cases there are predominant genera, which vary in proportion. The different environmental conditions from which the microorganism groups were extracted allow the development of associations of this nature. Among the predominant genera in all samples are Lactobacillus, Prevotella and genera of the family Acetobacteraceae.
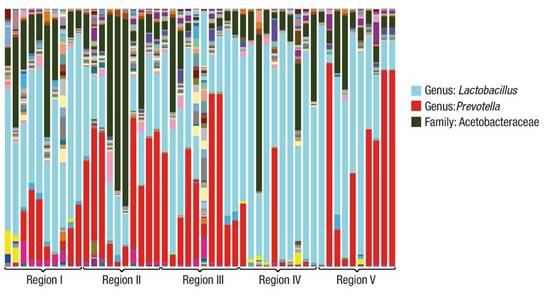
Figure 1 Variation of genera found in soil samples of the cloud forest ecosystem in the Sierra Madre del Sur in the state of Oaxaca.
Figure 2 shows the disappearance rate of pollutants up to concentrations of the order of 10 mg·L-1 in a period of 10 h, which was stable until the end of the experiment (120 h). The growth of the microorganisms was of the order of 1 x 1017, with the maximum being 1.2 x 1018. Initially, there is a direct dependence between the presence of pollutants and the increase in the number of microorganisms. It is possible that the maintenance in the number of microorganisms, subsequent to the drastic decrease in the concentration of pollutants, is due to the formation of the biofilm, which has a favorable environment to sustain the viability of the microorganisms present in the system.
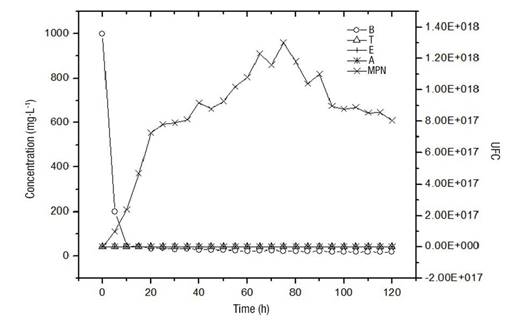
Figure 2 Disappearance rate of pollutants (B = benzene, T = toluene, E = ethylbenzene and A = anthracene) against microbial growth (MPN = Most Probable Number) in a packed-bed bioreactor.
Biofilm formation implies that several extracytoplasmic proteins interact with the abiotic surface and osmolarity of the medium (O'Toole & Kolter, 1998). The medium and conditions in which the development of the biofilm was promoted, and the change in the substrate from glucose to the pollutant mixture, may have activated the signal for the maintenance of bacterial viability. This signal, product of the change in environmental stimulus, translates into the expression of a mutant transposon as reported in the literature (Monteiro et al., 2007; O'Toole & Kolter, 1998; Wu, Yeh, Lu, Lin, & Chang, 2008).
The bacterial physiology required for the degradation of pollutants follows multiple mechanisms, including the activation of the cytochrome P450 enzyme pathway, which catalyzes the biotransformation of xenobiotics, which is a crucial step in this metabolic process (Huang, Nemati, Hill, & Headley, 2012; Monteiro et al., 2007; Janikowski, Velicogna, Punt, & Daugulis, 2002).
The change undergone by microorganisms in the nutritional aspect is the first significant stimulus, where there is no glucose and predominant or total presence of polluting substrates. This change is evident in the production of extracellular polysaccharides to form the bacterial biofilm (Figure 3), which have a structural function and act as surfactant agents (Ławniczak, Kaczorek, & Olszanowski, 2011).
Characterization of the biofilm by next generation sequencing shows the final behavior and establishment of communities and interactions along the length of the bioreactor. Figure 4 shows that the microbiological distribution had an important variation, despite maintaining constant temperature and oxygen saturation during the experiment, which are key factors in microbial aerobic metabolism. The distribution by sections in the bioreactor allowed establishing that the genera Bacteroides, Prevotella and Lactobacillus increased their presence up to 50 %.
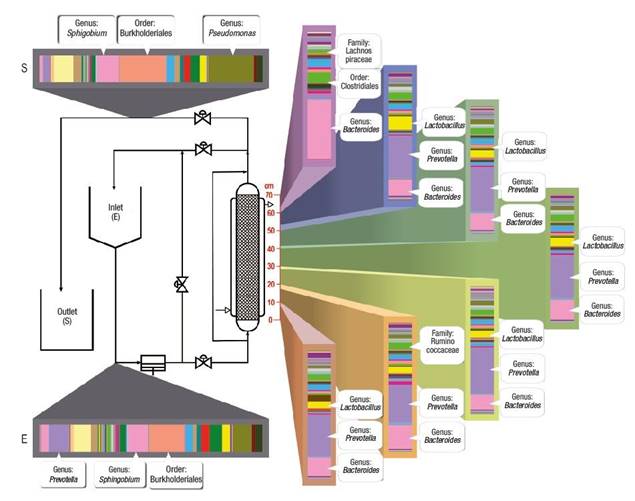
Figure 4 Variation of genera found along the bioreactor during the biodegradation process of aromatic hydrocarbons.
The degree of mineralization reached in the process is shown in Figure 5. The initial TOC corresponds to the amount of pollutants present in the solution. During the process, the IC increases due to the metabolic transformation that the microorganisms perform; however, at the end of the process, the sum of the TOC and the IC do not correspond to the initial TOC. The difference is the loss in the form of CO2, the waste product of the aerobic metabolism of microorganisms. During this process, the mineralization reached is 91 %. An additional stimulus such as the addition of a surfactant to the medium or a substrate favoring co-metabolism could increase the degree of mineralization.
Conclusions
The biodiversity found in the cloud forest archipelagos in the Loxicha region is key to ensuring ecosystem services, so it is important to undertake explorations to evaluate the use of these bacterial microbiomes. The genetic diversity of the cloud forest ecosystem in the state of Oaxaca and the balance achieved are due to the proportion of microorganisms involved, with Lactobacillus, Prevotella and genera of the family Acetobacteraceae prevailing. These microorganisms showed degrading activity on aromatic compounds (benzene, toluene, ethylbenzene and anthracene) that constitute hydrocarbon fuels, until their mineralization.











 text in
text in