Introduction
The p53 suppressor gene is located on chromosome 17p13, contains 11 exons with 20 kilobases, and encodes a nuclear phosphoprotein of 393 amino acids [1]. This gene plays a crucial role in preventing genetic mutations and maintaining genome integrity besides regulating the cell cycle, apoptosis, and cell differentiation, among other biological functions [2,3]. For this reason, p53 is known as “the guardian of the genome” [4]. The p53 mutation is one of the most frequent genetic alterations in human cancer, and more than 50% of cancers are related to this mutation [5]. When the p53 gene mutates it adopts oncogenic activities that initiate and promote tumorigenesis, progression, metastasis, and drug resistance [6]. In addition, this gene mutation allows diagnosis in early stages, prognostic and risk of recurrence evaluations, and determining the response of tumors to therapy [7,8]. Therefore, highly sensitive and reliable detection of the mutant p53 gene is essential for detecting, preventing, and treating cancer [6].
Several techniques have been developed to evaluate gene mutations such as Enzyme-linked Immunosorbent Assay (ELISA), electrophoretic immunoassays, mass spectrometry-based on proteomics, Radioimmunoassay (RIA), High-Performance Liquid Chromatography (HPLC), and Western blot or electron transfer, however, they use expensive materials and instrumentation and require well-trained personnel for their operation. On the other hand, biosensors have been implemented due to their accurate and specific detection of cancer biomarkers that had helped in early diagnosis and monitoring of disease progression [9,10]. A biosensor integrates a bioreceptor and a transducer. Bioreceptors are biomolecules that can recognize specific analytes such as antibodies, enzymes, and aptamers. As for the transducers, they convert the bioreceptor-analyte interaction into a measurable signal and include mass-based, optical, thermal, acoustic, piezoelectric, and electrochemical types [11]. The latter transforms the interaction between the biochemical receptor and a target analyte into an electrical signal like the voltage, current, or impedance. Sometimes these biosensors use an electrochemical probe to enhance the signal [10]. Electrochemical biosensors are highly selective in the determination of their counterparts because they can only recognize the bioreceptor-analyte event, this may lead to highly sensitive devices capable of detecting lower concentrations down to attomolar magnitudes (10-18) and with a wide dynamic linear range [12]. They also have the advantage of simple design, portability, easy handling, can be easily miniaturized, and are used for point-of-care testing [9,13]. In this sense, DNA-based biosensors (or genosensors) have generated considerable interest due to their fast, simple, and cheap detection of genetic diseases [1]. These can identify specific DNA sequences and their mutations due to their high sensitivity and specificity [9]. They use immobilized DNA as a diagnostic element and measure changes in current or resistance created by DNA hybridization [8]. They can be measured by electrochemical methods, such as Cyclic Voltammetry (CV) [14]. This technique evaluates electrochemical behavior in two different ways: direct and indirect. Direct methods are associated with the intrinsic electrochemical activity of nucleotide bases such as guanine and adenine [15] and do not require labeling for identification. In contrast, through indirect methods, it is possible to monitor changes in the electrochemical response of external labels (or indicators) to detect DNA hybridization [16]. DNA indicators are electroactive molecules that intercalate between two adjacent base pairs of double-stranded DNA (dsDNA) for its labeling, and intercalators have the advantage of not requiring chemical modification of the DNA probes in the construction of the biosensor to confirm detection [17,13]. DNA intercalators include propidium iodide (PI), methylene blue (MB), Höechst 33258 (HC), ethidium bromide (EB), acridine orange (AO), benzo[a]pyrene (BaP), and doxorubicin (Dox) [17]. Doxorubicin is a DNA indicator that intercalates into the double-stranded DNA (or hybrid DNA) through hydrogen bonds between the guanine of one DNA strand and the guanine of the opposite strand. This interaction between doxorubicin and hybrid DNA allows for highly sensitive and selective electrochemical monitoring of the hybridization process by means of the changes in the current peaks in the direct current voltammograms [13,18]. In the past few years, self-assembled monolayers (SAMs) have been used to develop and construct biosensors that have allowed them to reach lower concentrations. This is due to the balanced intermolecular interactions, the binding of the head group to the substrate, the functionalization of the terminal groups, and their molecular and monolayer structure that act as a hydrophobic and hydrophilic molecular barrier [19,20]. The alkylthiol 11-mercaptoundecanoic acid (MUC) plays an important role in the construction of DNA biosensors. It works as a spacer that improves the hybridization reaction, minimize non-specific adsorption and allow parallel DNA orientation therefore, higher densities, more uniform monolayers and improves the lifetime of electrochemical DNA biosensors [19,21-24].
Due to the high sensitivity, simplicity, rapid response, low cost, and compatibility with microfabrication technologies, electrochemical methods have been implemented as a strategy for the detection of DNA sequences, especially to detect the mutant p53 gene frequently present in human cancer [14]. Therefore, our study aimed to develop an electrochemical genosensor to detect the 175p2 mutation of the p53 gene employing Cyclic Voltammetry (CV). Hence, a screen-printed gold electrode (SPGE) was modified with 11- mercaptoundecanoic acid and DNA probes to monitor the current change of the [Fe(CN)6]3-/4- redox process ([Fe(CN)6]3-↔[Fe(CN)6]4-) influenced by the hybridization reaction, as well as the changes produced by the interaction of doxorubicin with the double-stranded DNA.
Experimental
Material and methods
All oligonucleotide sequences from the 175p2 mutation described in Table 1 were obtained from T4oligo Company (Irapuato, Gto). The chemicals used in this work were purchased from Sigma Aldrich (St Louis, MO): ethylenediaminetetraacetic (EDTA), phosphate buffered saline pH 7.4 (PBS), 11- mercaptoundecanoic acid (MUC), Trizma® hydrochloride (Tris-Cl), doxorubicin (Dox); and from Merck: Potassium ferrocyanide ([Fe(CN)6]4-) and Potassium ferricyanide ([Fe(CN)6]3-). Ultrapure sodium dodecyl sulfate (SDS) was purchased from MP Biomedicals, Inc. (Solon, OH). Ultrapure water from the Millipore Milli- Q system (resistivity 18.2 MΩcm) was used throughout this experiment.
Table 1 Oligonucleotide sequences from 175p2 mutation.
| Description | Sequence | Mer | |
| Probe | ssDNA | 5-SH-(CH2)6- CAT GAC GGA GGT TGT GAG GC-3 | 20 |
| Complementary target | cDNA | 5-GCC TCA CAA CCT CCG TCA TG-3 | 20 |
| Non-complementary Sequence 1 | nc1DNA | 5-CAT GAC GGG GGG TGT GAG GC-3 | 20 |
| Non-complementary Sequence 2 | nc2DNA | 5-GCC TCA CAA CCT CCG TCT AC-3 | 20 |
| Non-complementary Sequence 3 | nc3DNA | 5-CAG AGC ATC TAC GAC CAG AT-3 | 20 |
Assembly of the DNA biosensor
We used a screen-printed gold electrode (SPGE), which is a three-electrode system that has a working (Au), counter (Pt), and reference (Ag/AgCl) electrode. Since reactions usually take place near the electrode surface, the electrode material and its dimensions influence its ability to detect. Therefore, gold is an excellent material for electrochemical sensing, is conductive, is chemically stable, and does not corrode or tarnish [25].
For the construction of the DNA biosensor, we followed the methodology previously developed in our laboratory (Garcia-Melo et al.) [26]. The gold working electrode of the SPGE was incubated for 30 min at room temperature within a mixture of 10 µL 11-mercaptoundecanoic acid (1 µM) and 30 µL of 100 µM thiolated probe DNA of 175p2 mutation (ssDNA) diluted in PBS 0.01 M. This mixture was named MUCS. Afterward, the electrode was rinsed with 0.1% SDS (diluted in 0.01 M PBS, pH 7.4) and then with Milli-Q water to eliminate non-absorbed oligonucleotides. To detect the genetic alteration, 40 µL of 100 µM complementary target (cDNA) was incubated on the modified Au/MUCS for 30 min at room temperature. Then, the Au/MUCS/dsDNA was rinsed with 0.1% SDS (diluted in 0.01 M PBS, pH 7.4) and then with Milli-Q water to remove unhybridized oligonucleotides. After the hybridization assay, 10 µL of 20 µM doxorubicin (Dox) diluted in PBS (0.01 M, pH 7.4) was added to the Au/MUCS/dsDNA electrode for 20 min in the dark at room temperature. Previous studies show that the best effect of Dox is at 20 min. To remove the Dox molecules that did not interact with double-stranded DNA (dsDNA), the electrode was rinsed with Milli-Q water and then with PBS (0.01 M).
Electrochemical detection was performed with the Bioanalytical System BAS-100 electrochemistry workstation (West Lafayette, IN) with the Cyclic Voltammetry (CV) technique. Measurements were carried out following preliminary experiments [26] at each modified and unmodified electrode. We used 2.5 mM of the [Fe(CN)6]3-↔[Fe(CN)6]4- complex ([Fe(CN)6]3-/4- as the redox probe) diluted in 0.01 M PBS, pH 7.4 (total volume 150 µL) in the potential range of - 400 mV to + 500 mV, and a scan rate of 50 mV/s at room temperature. CV technique was used to investigate the oxidation and reduction process of the [Fe(CN)6] ¿3-/4- complex on the surface of the working electrode, and thus evaluate the current changes before and after each modification.
Sensitivity and optimization of experimental conditions
The sensitivity of the DNA biosensor was evaluated by incubating the Au/MUCS electrode with 40 μL of cDNA at different concentrations: 1fM, 10 fM, 100 fM, 1 pM, 10 pM, and 100 pM; for 30 min at room temperature. The rising process was implemented as described above. In the last step, Dox was added at each Au/MUCS/dsDNA as in the previous Dox protocol to assess its detection ability. To maximize DNA biosensor analysis performance, following the assembly of the DNA biosensor procedure, the hybridization temperature was evaluated. The Au/MUCS electrode was incubated with 40 µL of 100 µM cDNA at different temperatures for 30 min: 20°, 37°, 40°, 45°, 50°, and 55°C; using the US autoflow automatic CO2 (NU-4750) incubator (NuAire, Plymouth, MN). Then, the hybridization time was varied: 30, 40, 50, and 60 min; using 40 µL of 100 µM cDNA in the Au/MUCS at room temperature. Finally, since a real sample contains complementary and non-complementary DNA, the selectivity of the electrode was determined by carrying on the hybridization process of MUCS with the sequences: cDNA, and non-complementary (nc1DNA, nc2DNA, and nc3DNA) for 30 min of incubation time at room temperature. The CV technique was carried out following the protocol previously described, along with the presence and absence of Dox.
We used a new Au/MUCS electrode for each variation of concentration, hybridization temperature, hybridization time, and selectivity analysis.
Results and discussion
Electrochemical detection
To evaluate step by step the DNA biosensor construction strategy, we employed the CV technique on the unmodified and modified electrodes with 2.5 mM of the [Fe(CN)6]3-/4- redox complex diluted in PBS (pH 7.4) as the electrochemical probe. Fig. 1 (a) shows the electron transfer on the bare gold electrode surface for the reversible reaction: [Fe(CN)6]3- ↔ [Fe(CN)6]4-. When we scanned the potential from -400 mV to 500 mV through the electrode surface, the [Fe(CN)6]3-/4- complex is oxidized and the current increases until it reaches its maximum current. At the maximum anodic peak current, the volume of the solution containing the oxidized complex increases, which slows down the transport of ions between the bulk solution and the electrode, resulting in less current. Then, the complex is reduced as the potential becomes more negative, resulting in a decrease in the current [27]. In this way, we can observe the reversibility of the redox reaction [Fe(CN)6]3- ↔ [Fe(CN)6]4 on the gold electrode. It is well-known the [Fe(CN)6]3-/4- behavior in the potential range we are exploring in a gold electrode, for this reason, we established the Au voltammogram as a reference to compare the anodic peak current between each modification. We registered the maximum anodic peak current of the bare Au electrode (working electrode) in 77 µA at 160 mV (Fig. 1 (a)). These results demonstrate the efficient process of the electron transfer of [Fe(CN)6]3-/4-, a similar outcome was obtained in previous experiments [26].
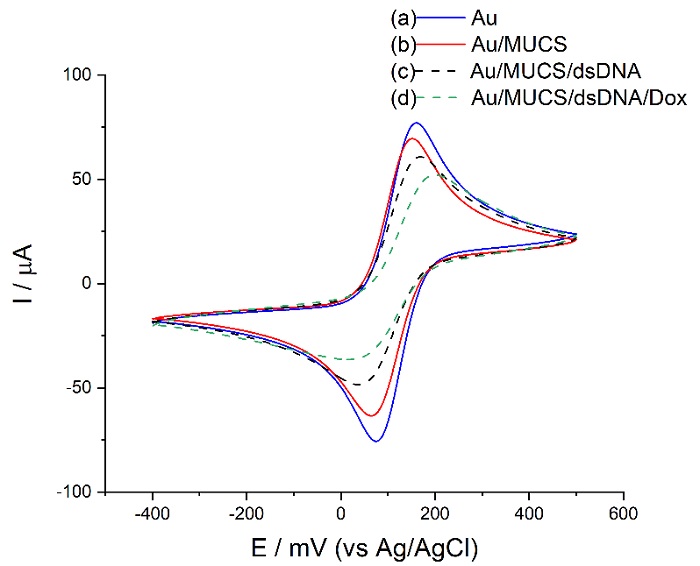
Fig 1 Cyclic voltammograms of unmodified Au electrode (a), modified Au/MUCS (b), Au/MUCS/dsDNA (c), Au/MUCS/dsDNA/Dox (d) performed in 0.01 M PBS (pH 7.4) containing 150 µL of the redox reaction 2.5 mM [Fe(CN)6]3-/4- at a scan rate of 50 mV/s.
Fig. 1 (b) shows the Au electrode modified with MUCS (self-assembled monolayer with ssDNA and MUC) and its maximum anodic peak in 69.46 µA at a potential of 152 mV. The decrease in the current of the [Fe(CN)6]3-/4- redox peak is due to the thiolated monolayer formation of MUC and ssDNA on the surface of the working electrode, where the negative charge of the phosphate backbone of thiolated DNA contributed to the repulsion of [Fe(CN)6]3-/4- ions on the surface of the electrode [28]. Besides, longer alkyl groups like 11- mercaptoundecanoic acid form more organized self-assembled monolayers that are free of defects and better block the access of the redox molecule on the electrode surface. For this reason, this alkyl molecule is used as an insulating material to block certain electrochemical reactions [29]. Additionally, MUC allows DNA probe flexibility, necessary for effective hybridization reaction, and after base pairing, it allows the formation of a rigid double-stranded DNA [30]. Moreover, according to topographical evaluations carried out in previous studies with Atomic Force Microscopy (AFM), when MUCS monolayer is created on the surface of the electrode, the genetic material adopts an organized and packed structure that does not allow the effect of pinhole from happening, and neither the electron transfer, as shown in the decrease in the current [31]. These results confirm that the DNA probe was successfully immobilized on the electrode surface.
It is worth mentioning, the current value of Au/MUCS represents 0% of hybridization and is the reference in the detection process of complementary DNA.
For the Au/MUCS/dsDNA electrode (Fig. 1 (c)), where the hybridization reaction took place, the peak current decreased by 18.36% IRelative compared to Au/MUCS. It is attributed to the contribution of the increasing molecular material of dsDNA (double-stranded DNA) in the electrode surface and the formation of a more compact adlayer which physically blocks access of the [Fe(CN)6]3-/4- ions to the surface of the electrode. Also, the increase of negative charge during DNA hybridization may be due to the double-stranded formation that creates more electrostatic repulsion force between the redox probe and the electrode surface, as reported in previous literature [28,32,33]. Using Dox (Fig. 1 (d)), peak current decreases by 22.94% IRelative concerning dsDNA, or 41.29% IRelative compared to Au/MUCS, which reflects a significant improvement in the sensitivity of the electrode. These changes in the current indicate intercalation between dsDNA and Dox on the electrode surface [34]. Dox intercalation on the nucleotide bases of the dsDNA was reported in some investigations [35- 37]. Moreover, in previous work, we evaluated different intercalating agents (PI, MB, HC, EB, AO, and Dox) in an electrochemical APC genosensor. Based on the results, we selected Dox since it showed a stronger capacity to interact with dsDNA and thus, indicated the hybridization process [26].
Doxorubicin in combination with the double-stranded DNA (dsDNA) affects the charge of the electrode surface and thus the transport of the redox probe [34]. This could be due to the changes in the charge distribution during the coating and the influence of DNA molecules over the redox molecules that creates an electrostatic repulsion between the dsDNA, Dox and [Fe(CN)6]3-/4- ions. Therefore, the redox probe struggles to pass through the DNA layer and access the electrode surface to exchange electrons [34,38]. Based on the previous statement, decrease in the peak current for Fig. 1 (d) is due to Dox binding to dsDNA that forms a larger and bulkier slowly diffusing structure [39]. In this sense, Kulikova et al. [40] mention that the changes in the current caused by Dox depend on the amount of DNA detected. On the other hand, our results show that the incubation time with Dox is sufficient to indicate the detection process of the double-stranded DNA, as reported by Hassani Moghadam et al. [41] where they confirmed that in 20 minutes Dox intercalates the double- stranded DNA. Moreover, Ma et al. [42] obtained similar results to ours. They used Dox as an intercalating agent and observed that 20 minutes is the ideal action time because after this period, no further adsorption will occur as they are limited by the DNA structure.
Our results allow us to suggest that Dox was intercalated in the cytosine and guanine base pairs of dsDNA in such a way that Dox forms conventional O-HO and O-HN hydrogen bonds with the DNA. Also, that there is an intermolecular interaction between the hydrogen atom of the hydroxyl group of the Dox agent resulting in an enhanced signal for the application and detection of our device [41,43,44].
Analytical electrochemical response
Using the CV technique, we evaluated the electrochemical activity of the genosensor at different cDNA concentrations: 1fM to 100 pM. The experiment was carried out at room temperature with an incubation time of 30 min. We used a new Au/MUCS electrode per evaluation. Additionally, we added Dox to improve the sensitivity of the detection of the hybridization reaction. Fig. 2 represents the maximum anodic peaks of the voltammograms with their error bar obtained in the concentration range previously evaluated. These results indicate a decrease from 58.41 to 40.59 µA for 1fM to 100 pM, respectively ((a), blue bar). Concerning Dox, a decrease from 53.83 to 38.98 µA was shown for the same concentration range ((b), red bar respectively). The results obtained using Dox may be related to its ability to intercalate on the double-stranded DNA, which causes a decrease in the current and an increase in the detection capacity. Similarly, Malanina et al. [45] indicate that this effect is due to the compaction of the superficial layers after the interaction with Dox, changing the spatial configuration of the biomolecules and increasing the negative charges on the electrode surface. As regards the error bars in figures 2-5, these values were obtained as a statistical result from Anova and Student's t-test by repeating the experiment five times.
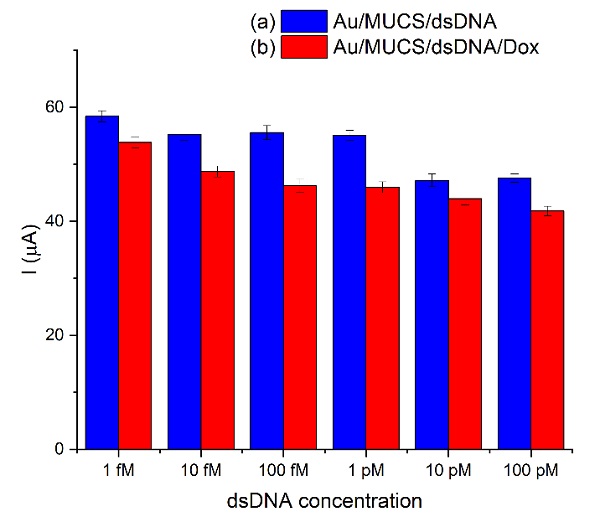
Fig 2 Maximum anodic peaks registered in the Au/MUCS electrode incubated at different concentrations of cDNA from 1 fM to 100 pM (a), and in the presence of Dox (b).
To calculate the limit of detection (LOD) of the device, we used the 3 x Sb criterion, where Sb is the standard deviation of the blank measurements which were fitted to the linear portion of their respective equation (the linear correlation between the logarithm of the cDNA concentration and the percentage of the relative current (% IRelative). The linear regression coefficient for the double-stranded DNA detection was R2= 0.91795 (Fig. 3 (A)). The LOD calculated was 2.20 x 10-15 M and the equation of the line was y= 67.64+3.53 log [dsDNA (M)] according to the method of Garcia-Melo et al. [26]. Fig. 3 (B) shows a better linear regression coefficient (R2= 0.97203) for dsDNA/Dox and a LOD of 1.75 x 10-16 M with an equation of y= 71.38+3.11 log [dsDNA/Dox (M)]. With Dox, the device shows a better linearity, and a lower LOD which means that the device can detect lower concentrations of the mutation with a better sensitivity than without Dox. The use of electroactive molecules leads to a more specific and sensitive detection because these molecules only interact with the dsDNA by intercalation, and this interaction is observed as a reduced signal due to a diminution of the diffusion coefficient of the electrode surface [46]. Therefore, we can conclude that the performance of our genosensor improved after the intercalation of Dox molecules into the double-stranded DNA.
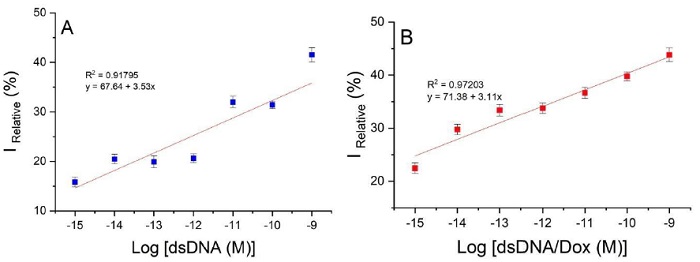
Fig 3 Calibration curve at different concentrations of log [cDNA (M)] (10-15 to 10-9 M) for Au/MUCS/dsDNA (A) and Au/MUCS/dsDNA/Dox (B).
Although all the components of the coating have been previously tested, their combination resulted in a remarkable increase in the sensitivity of the DNA sensor towards the model intercalator (doxorubicin). Doxorubicin intercalation influenced both the charge distribution and the volume of the biopolymer. Furthermore, it could promote changes in the aggregation of the reagents at the electrode interface [13].
When comparing these results with other devices for detection of the p53 gene, we could conclude that our genosensor has one of the highest efficiencies than more sensors today available. Table 2 shows the detection range and limit of detection of several devices for the p53 gene detection. It also shows that some authors employed complex characterization techniques like Surface plasmon resonance. Although this technique has high sensitivity, faster sample throughput and doesn’t need any radioactive or fluorescent labels; this technique needs a meticulously control experiments design for a reliable detection and to ensure the binding to the electrode chip surface. The immobilized probe may not maintain its native configuration upon immobilization on the surface, and its orientation may sterically hinder the analyte binding. Moreover, this technique employs expensive sensor chips and instrumentation [47].
Table 2 Analytical comparison of several devices for detection of the p53 gene.
| Electrode | Technique | Detection range | Limit of detection | Ref. |
| MOSFET/Au | Surface plasmon resonance | N/A | N/A | [48] |
| Au NPs @ PDA @ QDs | Electrochemiluminescence | 100 – 1.5 x 104 pM | 30 pM | [49] |
| PNA / nano-Au / Ch / GCE | Electrochemical | 50 – 9.6 x 104 pM | 18 pM | [50] |
| Streptavidin magnespheres paramagnetic particles | Electrochemiluminescence | 5 x 10-5 – 0.1 pM | 2 x 10-5 pM | [51] |
| Au-PANI (Polyaniline) | Electrochemical | 0.001 – 100 pM | 5 x 10-4 pM | [52] |
| nano-Au/Ch/GCE | Electrochemical | 50 – 9.6 x 104 pM | 18 pM | [53] |
| Au-nanorods | Surface plasmon resonance | 1 x 10−4 – 1 x 105 pM | 11.47 x 10-3 pM | [54] |
| Ag nanoparticle array | Surface plasmon resonance | N/A | N/A | [55] |
| Carbon paste electrode/PNA | Chronopotentiometric | 4 x 104 –1.2 x 106 pM | 4 x 104 pM | [56] |
| Au-MWNTs-PA6-PPy | Electrochemical | 0.1 – 100 pM | 0.05 pM | [57] |
| Au-PNA-dsDNA-MB | Electrochemical | 1000 – 1 x 107 pM | 682 pM | [16] |
| dsDNA/MCH - AgNPs/BDT | Electrochemical | 0.1 – 100 pM | 0.1 pM | [18] |
| SAM-Au/Fc | Electrochemical | N/A | 2.2 pM | [58] |
| AuNPs/GO | Electrochemical | 0.2 – 200 pM | 0.03 pM | [5] |
| CuNPs/DNA | Fluorescence | 100 – 2 x 105 pM | 52 pM | [59] |
| AuNP-SPCE-HRP-p53392Ab2-GO | Electrochemical | 20 – 2000 pM | 10 pM | [60] |
| AuNP-SPCE | Electrochemical | 2000 – 5 x 104 pM | 50 pM | [61] |
| GO–CHI/SPCE | Electrochemical | 200 – 1 x 104 pM | 100 pM | [62] |
| Au/MCH/PNA/IC | Electrochemical | 10 – 2 x 106 pM | 4.31 pM | [63] |
| Au/MWNTs–Ru(bpy)3 2+-PPy | Electrochemiluminescence | 0.2 – 200 pM | 0.1 pM | [64] |
| Au/MUCS/dsDNA | Electrochemical | 0.001 – 100 pM | 2.2 x 10-3 pM | Our |
| Au/MUCS/dsDNA/Dox | Electrochemical | 0.001 – 100 pM | 175 x 10-6 pM | Our |
Some of the electrodes on Table 2 have complex chemical surface modifications that employs expensive reagents in order to detect with specificity and sensitivity. But in ours, we don’t have to modify the structure of the electrode surface in order to have a good performance. We create compact layers with the self- assembled monolayers (SAMs) modification technique that allows the correct, stable, and free of defect immobilization of the DNA probe. With SAMs, we modified physically the surface of the electrode without changing the chemical properties of the electrode. Our technique is simple, reproducible, and efficient, and that has allowed us to be on the list best selective and sensitive biosensors for the p53 gene detection.
Optimization of the experimental conditions
To maximize the performance of our DNA biosensor, we optimized the experimental conditions by the variation of temperature and hybridization time. The hybridization temperature is a key factor that affects the performance of the genosensor. Elevated temperatures are usually used to improve the hybridization specificity; at these temperatures, DNA probes will have a better ability to distinguish and bind their complementary parts according to their affinity properties [65]. Therefore, we incubated Au/MUCS at temperatures of 20, 30, 37, 40, 45, 50, and 55°C, for 30 min with and without Dox. Fig. 4 (A) (blue bar) shows that the highest influence of temperature on the hybridization reaction is at 20 and 55°C with a current of 41.51% IRelative and 44.74% IRelative, respectively. These results indicate that the genosensor can work with the highest efficiency at both temperatures. However, when Dox was used, the sensitivity increased for all temperature ranges, but at 20°C, the genosensor showed the best response with about 67% IRelative (Fig. 4 (A), red bar). According to several reports, temperatures 5°C below the sequence’s melting point are optimum for hybridization reaction [66], for this reason at 55°C there is no impediment to base-pairing. The genosensor behaves similarly when Dox is used at room temperature (20°C). Therefore, the action of Dox is not temperature-dependent, and it is possible to work at 20°C and detect the hybridization reaction. Hence, it is no longer necessary to incubate at elevated temperatures to be able to have 100% hybridization. Castaneda et al. [67] observed similar behavior when they used methylene blue as an intercalating agent, the molecule decreased the current and exhibited its greatest effect at room temperature.
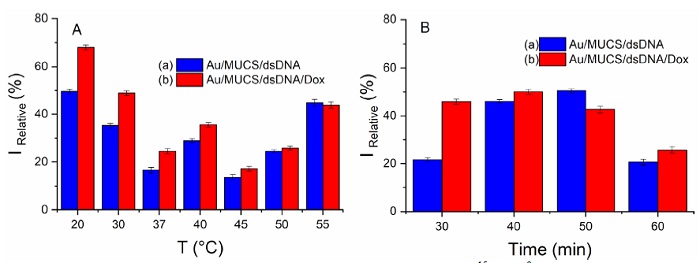
Fig 4 Calibration curve at different concentrations of log [cDNA (M)] (10-15 to 10-9 M) for Au/MUCS/dsDNA (A) and Au/MUCS/dsDNA/Dox (B).
On the other hand, the optimization of the hybridization time was carried out by incubating 100 μM of cDNA on the Au/MUCS electrode at different incubation times of 30, 40, 50, and 60 min at room temperature. A different Au/MUCS electrode was used for each incubation time. Fig. 4 (B) (blue bar) represents the maximum anodic peaks of [Fe(CN)6]3-/4- for each hybridization time for Au/MUCS/dsDNA. The genosensor shows the highest response in 40 and 50 min, with 46.09 and 50.51% IRelative, respectively. However, the behavior of the DNA biosensor changed when we Dox added. The DNA biosensor showed a greater response at 30 and 40 min; with a current of 45.95 and 50.04% IRelative, respectively. When we compared the data shown in Fig. 4 (B), we observed that at 40 min the device had an increase of 107.17% compared to Au/MUCS/dsDNA, while at 30 min, the current increased 212.33%. Based on these results, 30 min was selected as the optimal hybridization time.
Selectivity study
The selectivity of the genosensor was determined by carrying on the hybridization process with a mixture of complementary DNA target (cDNA) and non-complementary sequences (nc1DNA nc2DNA and nc3DNA) to emulate a real sample (see Table 1). This mixture was incubated on the genosensors surface for 30 min, at room temperature. The results presented in Fig. 5 indicate a higher current change in the detection of the complementary DNA sequence in comparison with the non-complementary ones; the maximum anodic peak of the Au/MUCS/dsDNA electrode decreased 23.79 µA (red bar) compared to Au/MUCS (black bar). Then, we observed a decrease of 32.75 µA while using Dox with the complementary target (blue bar) but an imperceptible difference with the non-complementary sequences.
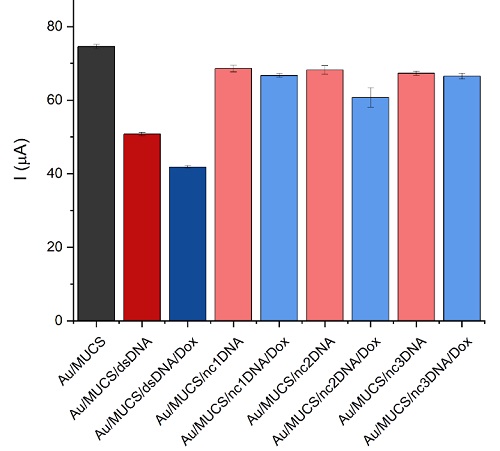
Fig 5 DNA biosensor response in presence of target and non-complementary DNA probes (nc1DNA nc2DNA and nc3DNA).
Hybridization with no matching sequences often results in a slight decrease in current, which means that not all bases matched with the immobilized probe (Au/MUCS). As observed in the hybridization process, when the complementary sequence pairs with the 175p2 immobilized DNA probe (ssDNA), a decrease in the anodic peak current can be observed. This can be attributed to 1) an increase in the negative charge on the electrode surface due to the formation of dsDNA that is negatively charged and 2) an increase of material on the electrode surface that physically blocks the [Fe(CN)6]3-/4- ions from circulating through the electrode surface [28,32,33]. However, when non-complementary sequences interact with the ssDNA there is a slight change in the maximum peak current. During the hybridization assay with the non-complementary sequence, certain kinks may form due to a mismatched sequence. In these spaces, where no base pairing occurred, the ion transfer takes place, and we observed a slight change in the peak current indicating that certain bases of the sequence match the non-complementary ones and the majority are mismatched [68]. Under these conditions, no complete formation of dsDNA results in a slight decrease in current, almost imperceptible because the negative charges of the dsDNA backbone are insufficient to prevent the ion transfer of [Fe(CN)6]3-/4- ions. Therefore, no decrease in current can be associated with no complete hybridization. However, when we added Dox to the non- complementary sequences, it influenced a slight current decrease because it was intercalated between the sequences where there was no mismatch.
These results are attributed to the intercalation mechanism of Dox, which has a higher affinity for double-stranded DNA than single-stranded oligonucleotides. Therefore, the nanobiosensor has the ability to distinguish between the target DNA, which forms a complete double strand with the DNA probe, and non- specific oligonucleotides which cannot form an incomplete double-stranded structure [69].
Our device is selective because it only showed a significant current decrease in the presence of the complementary strand, with and without Dox. It was able to identify between different sequences and only interact with its complementary part. In general, our results confirm the ability of Dox to interact exclusively with fully complementary sequences. In the same way, nc2DNA showed a greater effect after adding Dox compared to nc1DNA and nc2DNA. That can be attributed to the similarity between nc2DNA and cDNA; nc2DNA can match some of the bases of the immobilized probe, but the current decrease shown is not enough to assess a detection. Finally, the use of intercalating molecules such as doxorubicin increase the detection capacity of electrochemical biosensors. Therefore, the selectivity and sensitivity of our genosensor has been improved by using Dox as indicator of the hybridization reaction.
Conclusion
To summarize, we employed a simple structuring to develop an electrochemical genosensor with high sensitivity and selectivity, capable of detecting the 175p2 mutation of the p53 gene. The genosensor was based on the immobilization of MUCS adlayer at the SPGE surface and the use of Dox as an indicator of the hybridization reaction. Its simple design managed to reach the detection limit of 175 aM. The best performance was obtained after 30 minutes of hybridization reaction at room temperature in the presence of Dox. This electrochemical genosensor was successfully and efficiently applied to non-complementary sequences, and it demonstrates its ability to detect just its complementary target, demonstrating that it has significant potential for selective and sensitive detection of the 175p2 mutation of the p53 gene. Therefore, this electrochemical genosensor would be an excellent tool to identify disorders related to this genetic alteration and be considered in the preclinical area. The method discussed in the present investigation can be adapted for the investigation of other mutations of several types of diseases, in addition to being applied to clinical diagnosis and biomedical research.











 nueva página del texto (beta)
nueva página del texto (beta)


