Introduction
Marigold (Tagetes erecta L.) (Asteraceae) is endemic to Mexico and is commonly used as an ornamental and medicinal plant [1]. Marigold flowers have been used as a source of natural dyes, mainly to colour chickens’ skin and egg yolks, due to their high carotenoid content [2]. Lutein (C40H50O2) is the main pigment in yellow-orange flowers and represents up to 90 % of the total carotenoid content. Antioxidant and antimicrobial (against Fusarium oxysporum and Trichophyton mentagrophytes) activities have been reported for the essential oils from flowers and leaves of T. erecta L., respectively [3,4]. Progress in transforming different plant species has been achieved mainly by using different strains of Agrobacterium tumefaciens, combined with an improvement of regeneration techniques from specific explants. Stable transformation by particle bombardment with the uidA gene [5], transient transformation via Agrobacterium tumefaciens [6], stable transformation using A. tumefaciens strain LABA4404 containing the binary vector pBI121 [7] and overexpression of chrysanthemyl diphosphate synthase gene leads to the overproduction of pyrethrin [8] have been previously reported for T. erecta L. Here, we report for the second time a stable transformation protocol for obtaining transgenic plants of T. erecta L. var. Marvel Orange with the GUS reporter gene via A. tumefaciens.
Experimental
Plant material
Marigold (T. erecta L.) var. Marvel orange [Cat 8061]) seeds were obtained from BallTM (USA). Seeds were superficially disinfested with 20 mL 0.01 % (w/v) Captan solution and 0.03 % (w/v) Benlate (Benomyl) for 20 minutes, 5 % extran for 5 minutes, 80 % ethanol for 5 minutes and 2.5 % sodium hypochlorite for 30 minutes. Disinfested seeds were placed on MS medium (Phytotechnology Laboratories [Cat M571], pH 5.7, supplemented with 0.4 g·L-1 NH4NO3 and 3 % sucrose) without plant growth regulators. Germination was carried out under continuous light (40-50 m2 s mol-1).
Transformation procedure
Shoot-tips from 3-week-old seedlings were dissected and incubated with A. tumefaciens LBA4404 strain, previously transformed with the vector pCAMBIA 2301 through heat shock. This binary vector contains the neomycin phosphotransferase II gene (NPTII) that confers kanamycin resistance to transformed shoots and also carries the uidA gene, encoding a β-glucuronidase with a catalase intron (the presence of this intron ensures expression in eukaryotic cells and thus, preventing it in bacteria) that provides phenotypic selection of the transformed explants, through the characteristic blue colour, resulting from β-glucuronidase activity on X-GLUC in transformed plant tissues. Both genes are under the control of the CaMV35S promoter. A. tumefaciens LBA4404 strain harboring pCAMBIA 2301 was cultivated in 20 mL YEB medium (containing 100 mg·L-1 rifampicin, 50 mg·L-1 kanamycin and 100 mg·L-1 streptomycin) in the dark for 48 hours at 28 °C under constant agitation (200 rpm). After this period, 2 mL of bacterial suspension was re-inoculated in 40 mL YEB medium with the same antibiotics and then incubated for further 24 hours under the same conditions as before. Once this period was finished, 40 mL YEB medium with antibiotics and supplemented with 100 µM acetosyringone were added to the flask and then incubated for 3 other hours until an OD600= 0.6 was obtained. Seventy-five shoot tips were infiltrated using the mentioned bacterial suspension by applying a vacuum (40 cm Hg) for 15 minutes, then in order to eliminate any bacterial excess, tissues were blotted on sterile filter paper and placed in Petri dishes with MS semi-solid medium with 10 µM IAA and 70 µM BA at 25 °C in darkness. After 5 days of co-cultivation, the explants were rinsed with liquid MS medium with 100 mg·L-1 cefotaxime and placed on MS selective medium (MSs), which contains 10 µM IAA and 70 µM BA supplemented with 100 mg·L-1 cefotaxime and 50 mg·L-1 kanamycin. The explants were subcultivated every 30 days on MS and buds-shoots were obtained after 2 months. When the differentiated shoots reached a length of 3 cm, they were excised and placed in a flask with a semisolid MS medium free of growth regulators to promote rooting.
GUS histochemical stain
For histochemical GUS staining, shoot-tips recently transformed, as well as shoots, leaves, and flowers, were infiltrated under vacuum in X-GLUC (5-bromo- 4-chloro-3-indolyl-β-D-glucuronide) buffer [9] for 15 minutes, and then, incubated for 24 hours at 37 °C in darkness. The explants were thoroughly rinsed with a methanol:acetone solution (3:1) and then placed in a water:glycerol solution (1:1).
PCR analysis
To demonstrate NPTII genomic integration in the genome of T. erecta L., DNA was isolated using the procedure reported by [10] and PCR was performed to amplify a 600 bp fragment of the NPTII gene. PCR amplification profile for NPTII included an initial denaturation step at 94 °C for 3 min, 30 cycles of denaturation at 94 °C for 60 s, annealing at 57 °C for 45 s and extension at 72 °C for 120 s, with a final extension for 5 min at 72 °C. As a positive control, binary vector pCAMBIA2301 was used as a DNA template. DNA extracted from non-transformed plants was used as a template in the negative control. The amplified product was visualized on 1 % agarose gel stained with 1 µg∙mL-1 ethidium bromide.
Results and discussion
Previously, [6] had reported T. erecta L. transient transformation of several explants (shoot-tips, leaf primordia, hypocotyls, and roots) with gus reporter gene, thus showing the species’ susceptibility to genetic transformation via A. tumefaciens, but without regeneration of transformed plantlets. [5] Reported stable transformation of T. erecta L. using particle bombardment through the presence of blue spots on leaf explants but did not show positive histochemical staining of regenerated shoots and reported solely the number of insertion events in their regenerated shoots. More recently, [7] reported the stable transformation using A. tumefaciens strain LBA4404 containing the binary vector pBI121.
In this work, shoot-tips obtained from three-week old seedlings were infected with A. tumefaciens strain LBA4404, harboring the binary vector pCAMBIA 2301, and co-cultivated for 5 days. After this period, the histochemical assay showed positive staining in all explants (Fig. 1(B)). After three months, shoots could be regenerated, and after performing the histochemical assay again, explants showed complete staining of the stem and primary and secondary veins of the leaflets (Fig. 1, (E) and (G)).

Fig. 1 Stable expression in shoot-tip, shoot, leaf, and flower from seedlings regenerated from apical meristem of T. erecta L. var. Mavel orange via A. tumefaciens harboring pCAMBIA 2301 vector. (A) Untransformed shoot-tip (control); (B) shoot-tip transformed with pCAMBIA 2301 via A. tumefaciens after 5 days in MS regenerating medium (supplemented with 10 mM IAA and 70 mM BA); (C) untransformed, regenerated shoot (control); (D) regenerated shoot on medium with growth regulators (10 µM IAA and 70 µM BA) and antibiotics (50 mg L-1 kanamycin and 100 mg L-1 cefotaxime) showing positive GUS stain; e) transformed seedling after 3 months in MS medium without growth regulators; (F) leaf from untransformed, regenerated shoot (control); (G) leaf from regenerated shoot showing positive GUS stain; (H) flowers of Tagetes erecta L. var. Marvel orange obtained in vitro; (I) untransformed flower bleached with methanol:acetone solution (3:1) used as a control; (J) transformed flower showing positive GUS stain. Bars in a, b, f, g = 1 mm; in c, d, e, h, i, j = 1 cm.
In vitro flowering has been reported for several species of the Asteraceae family: Anthemis xylopoda [11], Vernonia cinerea [12], Pentanema indicum [13] and T. erecta L. During propagation of the potentially transformed T. erecta L. shoots in MS medium free of growth regulators (but supplemented with 100 mg·L-1 cefotaxime and 50 mg·L-1 kanamycin), flowering was observed in some cases (Fig. 1(H)). The ability to flower in vitro depends on numerous external, internal, physical, chemical, genetic, and environmental factors, which interact among themselves in panoply of ways [12]. GUS histochemical assay was performed in T. erecta L. flowers, and similar to shoot-tips, buds and leaflets, flowers presented an evenly distributed blue stain (Fig. 1(J)), thus demonstrating that the gene reporter (uidA) was functional in T. erecta L. flowers. A disadvantage of in vitro flowering is early senescence, since flowers that are not pollinated become infertile. All these plants died within a week after flowering.
Table 1 summarizes data obtained in this study in relation to shoot formation and transformation efficiencies. The shoot forming capacity index (SFC) was calculated according to [14], in order to determine the efficiency of a given treatment, as follows:
Table 1 SFC (14) = (SPC) X (SFE) / 100; IF (6) = number of GUS positive explants / total number of infected explants) x 100; TE (16) = (number of identified plants by PCR / number of inoculated explants) x 100.
| Shoots per callus (SPC) | Shoot forming explants (SFE) | Shoot forming capacity index (SFC) | Infection frequency (IF) (%) | Transformation efficiency (TE) (%) | |
| Transformed | 2.5 | 40 | 1 | 100 | 40 |
| Non-transformed | 2.66 | 15 | 0.4 | --- | --- |
Where SPC, the mean number of shoots per shoot apex-derived calluses, represents the ability of an explant to produce shoots, and SFE, the percentage of explants forming shoots, corresponds to the responsiveness of the tissue to the medium. In this work, SPC was 2.5, and SFE, 40 %, resulting in a SFC of one. Control explants showed lower SFC than transformed explants, as has been reported with transformed Brassica oleracea L. cotyledons [15]. Transformed T. erecta plantlets were placed on MS medium free of growth regulators (but supplemented with antibiotics) for rooting. After further 3-4 weeks, the seedlings generated roots (average length: 4-5 cm).
Here, the stable transformation of T. erecta L. var. Marvel Orange from shoot tips via A. tumefaciens has been obtained, with a higher transformation efficiency (40 %; Table 1), when compared to that achieved from leaf explant (16 %) using particle bombardment [5] but is lower that achieved (66 %) with hypocotyl explanst [7].
To verify the presence of NPTII gene in regenerated plants, PCR analyses were performed. DNA was obtained from leaves of regenerated plantlets, and after PCR, a 600 bp fragment could be detected in the gel (Fig. 2, lanes 1 to 8), thereby demonstrating that there was an NPTII transgene insertion. No DNA fragment was amplified when DNA extracted from non-transformed plantlets was used as a PCR template (Fig. 2, lane 9). [5] amplified a 617 bp fragment of the NPTII gene and through Southern blot demonstrated the presence of a single copy in the regenerated plants, thus the stable transformation of T. erecta L. In barley lines, which were transformed using A. tumefaciens via biolistic, [17] demonstrated that all of them integrated at least one copy of the transgene (luciferase) and in 60 % of these, there were eight or more integrated copies of the transgene. The insertion of multiple copies has been commonly associated to gene silencing [18]. One advantage of Agrobacterium-mediated transformation is that it facilitates the integration of a small number of copies of the transgene and thence, has a higher level of stability for the expression of desirable agronomic traits [19].
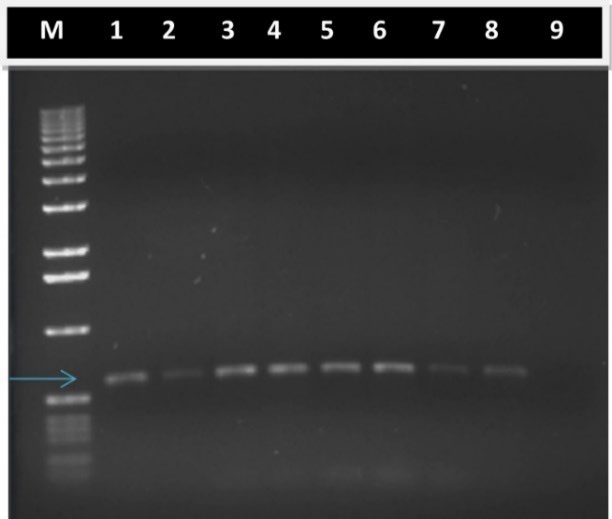
Fig. 2 Stable integration of transgene DNA into the genome of T. erecta L. var. Mavel orange transformed plantlets. Five microliter aliquots from PCR were fragmented by agarose electrophoresis and stained with ethidium bromide. Lane M. 1 kb DNA ladder as reference of fragment size; Lane 1. Positive control, DNA from binary vector pCAMBIA2301 was used as PCR template; Lanes 2-8. DNA extracted from transformed T. erecta L. plantlets was used as PCR template. Arrow indicates position of 600 bp; Lane 9. Negative control, DNA extracted from non-transformed T. erecta L. plantlets was used as PCR template.
Conclusions
Transformation of Tagetes erecta L. have been difficult. There are many factors involved, in sable transformation of this species, but from our experience on Tagetes erecta L., the variety is the key for successful. To the best our knowledge, this work is the second report of a successful stable transformation but using shoots tips of T. erecta L. plants with GUS reporter via A. tumefaciens. This transformation protocol can be used to introduce genes in T. erecta L., to manipulate the two isoprenoids pathways: mevalonate pathway (cytoplasmic) and MEP pathway (chloroplastic).
Reports of stable genetic transformation of T. erecta L. via A. tumefaciens are limited, so this work demonstrates the ability of A. tumefaciens to introduce foreign genes into shoot tips from T. erecta L., and the possibility to transform this specie with the gene DXS (CLA1) encoding the enzyme 1-deoxy-D-xylulose 5-phosphate synthase, which catalyzes the first limiting step in the MEP pathway, in order to be able to manipulate carotenoid biosynthesis.











 nueva página del texto (beta)
nueva página del texto (beta)


