Zea mays, commonly known as corn, is the most widely produced cereal crop worldwide, with wheat and rice following as close contenders (CIMMYT, 2019). It is worth noting that Mexico is considered the center of origin for corn (Matsuoka et al., 2002) and has the largest annual planted area and per capita consumption of this crop (García-López and Giraldo, 2021; Padrón et al., 2013). However, Mexico’s corn production and grain quality are often compromised by several fungal diseases, among which corn ear rot diseases are particularly prevalent (Moreno-Limón et al., 2011).
Fusarium fungi cause corn ear rot, which poses a significant limitation on the production and safety of corn worldwide. It is reported that most corn hybrids cultivated globally are susceptible to more than 10 Fusarium species causing ear rot (Mesterhazy et al., 2012). These species typically produce mycotoxins that affect not only animal health but also human health (Holf, 2020; Mielniczuk and Skwaryło-Bednarz, 2020). In Mexico, F. verticillioides is the most widely distributed and important species causing ear rot in the states of Guanajuato, Guerrero, Hidalgo, Jalisco, Puebla, and Nuevo Leon. Additionally, F. graminearum and F. subglutinans have also been reported to cause ear rot in the states of Mexico, Michoacan, and Yucatan (Zenteno-Zevada, 1963).
Biocontrol of plant diseases with antagonistic microorganisms is a useful strategy to reduce the use of pesticides in agriculture and lower production costs and environmental impact (Dimkić et al., 2022; Guzmán-Guzmán and Santoyo 2022; Luo et al., 2022; Singh et al., 2022). Previous studies have shown that exudates from the sclerotia of phytopathogenic fungi such as Sclerotium rolfsii and Sclerotinia sclerotiorum stimulate the growth of specific bacterial populations in the same agroecosystem with greater antifungal activity against these same fungi, compared to bacterial strains isolated from other ecological niches (Abdullah et al., 2008; Coley-Smith and Dickenson, 1971; Gilbert and Linderman, 1971; Hou et al., 2006). Bacterial populations that coexist in specific habitats with limited nutrients, such as sclerotia, promote the production of metabolites that increase their ability to compete against other microorganisms. Therefore, sclerotia of fungi are considered potential natural reservoirs of efficient antagonists for the biological control of phytopathogens (Hou et al., 2006; Zachow et al., 2011).
Claviceps gigantea, an ascomycete, causes the disease commonly known as “horse’s tooth” due to the shape of its sclerotia. This fungus produces several alkaloids, mainly ergoline, festuclavine, dihydroelymoclavine, chanoclavine, and elimoclavine, which are also toxic to both animals and humans (Agurell et al., 1963; Solano-Báez et al., 2018; Mielniczuk and Skwaryło-Bednarz, 2020; Bragg et al., 2017; Hof, 2020). C. gigantea is an endemic fungus limited geographically to locations in Mexico with altitudes above 1800 masl and relative humidity ≥ 60% (Fucikovsky and Moreno, 1976; Fuentes et al., 1964; Ullstrup, 1973). In the high valleys of Mexico (>2000 masl), maize is grown in environments with temperate climates, where ear rots caused by Fusarium spp. are common, often occurring simultaneously with C. gigantea, causing losses of up to 100% (CIMMYT, 2004). In this study, we hypothesize that C. gigantea sclerotia harbor bacterial populations with efficient antagonism against Fusarium species causing maize ear rot in Mexico. Our objectives were to: i) explore the bacterial populations associated with C. gigantea sclerotia in the State of Mexico, ii) evaluate the in vitro antagonism of these bacterial populations against Fusarium graminearum, F. subglutinans, and F. verticillioides, iii) molecularly identify the most efficient antagonists and characterize the in vitro production of their metabolites.
Materials and methods
Collecting Claviceps gigantea sclerotia
Sclerotia of C. gigantea were collected from six localities in the State of Mexico, which showed severe history under natural conditions of “horse tooth” disease and ear rot caused by Fusarium spp. (see Table 1 and Figure 1). For each locality, sclerotia were collected from C. gigantea-infected ears without symptoms of rot caused by Fusarium spp.
Table 1 Collection of C. gigantea sclerotia in localities of the State of Mexico.
| zLocalidad | Latitud (N) | Longitud (O) | Altitud (msnm) |
|---|---|---|---|
| Almoloya de Juárez | 19° 14´ 10” | 90° 42´ 07” | 2600 |
| Atlacomulco | 19° 43´ 37” | 99° 42´ 12” | 2700 |
| Calimaya | 19° 10´ 25” | 99° 32´ 12” | 2680 |
| Mina México | 19° 40´ 35” | 99° 40´ 10” | 2580 |
| Toluca | 18° 59´ 00” | 99° 40´ 58” | 2600 |
| Villa Victoria | 19° 26´ 00” | 100° 00´ 00” | 2570 |
z Sample size of 10 sclerotia per locality estimated with Cochran’s model (1982).
Isolation of bacteria from Claviceps gigantea sclerotia
Ten sclerotia of Claviceps gigantea were collected from each of the six locations. The sample size for the number of sclerotia was determined using the model described by Cochran (1982). The sclerotia (n=10) from each location were ground separately in a sterile mortar. One gram of ground sclerotia was diluted in 100 mL of sterile distilled water to perform serial dilutions up to 10-4. From each dilution, 100 µL was plated on Petri dishes with R2A culture medium (Difco) in triplicate and incubated at 28 °C for 24 h. Bacterial growth was quantified using the direct plate count method, and microbial density was expressed in Log10 CFU g-1 of sclerotium (Peng et al., 2009). A total of 129 bacterial isolates associated with sclerotia, exhibiting different colony morphology observed under a stereoscopic microscope (American Optical AO), were selected from the six sampled locations for further study of in vitro antagonism.
In vitro antagonism against Fusarium spp.
The in vitro antagonism of 129 bacterial strains was evaluated against three Fusarium species (F. graminearum, F. subglutinans, and F. verticillioides) responsible for ear rot in the sampled locations. The pathogenicity of these strains was experimentally verified in seedlings of three native populations of corn in the State of Mexico by inoculating the fungus in the substrate. The internal regions ITS of the rRNA 18S-5.8S and 5.8S-28S genes were molecularly identified through amplification, provided by Dr. Dolores Briones Reyes from the Graduate Program of Genetic Resources and Productivity of the Colegio de Postgraduados. In vitro antagonism was evaluated by dual confrontation on square Petri dishes (120 x 120 mm) with Waksman agar culture medium, which was selected from King’s B media, R2A, nutrient agar, and potato-dextrose agar, as it allowed optimal growth of both Fusarium species and bacterial isolates. The initial inoculation of Fusarium species strains was carried out by extension with an L-digralsky loop on the culture medium surface, and the inoculated Petri dishes were kept at room temperature for 60 min. Bacteria (n=129) were subsequently inoculated with a multi-point inoculator (Boekel®, 96-point microplate replicator) and incubated at 28 °C for 7 days. Bacterial strains that showed antagonism (inhibition halo of fungal growth) against one or more Fusarium species were selected. In another assay, antagonistic bacterial strains were individually inoculated by puncturing the bacterial mass with a sterile stick under the same experimental conditions as the previous assay. Only bacterial strains that showed antagonism by forming an inhibition halo ≥ 5mm of mycelial growth against one, two, or all three evaluated Fusarium species were selected. Daily observations were made, and the assay was repeated three times. Controls consisted of Petri dishes with growth of Fusarium species in the absence of antagonistic bacteria.
Qualitative detection of metabolite production
The antagonistic bacteria were qualitatively characterized for their ability to produce metabolites in vitro, such as indole-3-acetic acid, using soy tryptone broth (TSB) as the culture medium. Bacterial strains that produced a reddish coloration in the medium were considered positive (Frey-Klett et al., 2005), while siderophores were assessed using the universal medium chrome azurol S (CAS), with bacterial strains that evidenced a yellow halo around the colony being considered positive (Schwyn and Neilands, 1987). Lipolytic and proteolytic activities were evaluated using nutrient agar supplemented with Tween 80 and TSB medium supplemented with skimmed milk, respectively. Bacterial strains that evidenced an opaque and clear halo around the colony were considered positive for lipolytic and proteolytic activities (Hantsis-Zacharov and Halpern, 2007). Mineral phosphate solubilization was assessed using TCP medium, with bacterial strains that evidenced an opaque halo around the colony being considered positive (El-Yazeid et al., 2007). All evaluations were performed in triplicate.
Molecular identification of antagonistic bacteria
The DNA of the potential antagonists was obtained from individual colonies with 72 h of growth at 28 °C on Waksman agar medium using the PureLink Genomic DNA kit (Invitrogen Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocol. Partial amplification of the 16S rRNA gene was carried out using the primers 8F (5’ AGAGTTTGATCCTGGCTCAG 3’) and 1492R (3’ GGTTACCTTGTTACGACTT 5’) and PCR conditions described by Baker et al. (2003). The amplified products were sequenced (Sanger sequencing) at Macrogen Inc. (Seoul, Korea) (https://www.macrogen.com); the sequences were compared in the gene bank (GenBank) of the International Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/Blast) using the Blastn (Basic Local Alignment Search Tool) 2.2.29+ algorithm (Altschul et al., 1990).
Results
Bacterial population density in Claviceps gigantea sclerotia
The study revealed bacterial growth in R2A culture medium obtained from sclerotia at all sampling locations. Colony counting was performed to determine bacterial density in C. gigantea sclerotia, which varied from 2.023 to 2.397 Log10CFU g-1 of sclerotium across the six sampling sites (Table 2). Notably, the highest bacterial density was found in Atlacomulco lots (2.397) and the lowest in Villa Victoria (2.023) Log10CFU g-1 of sclerotium. These findings imply the presence of bacterial populations associated with C. gigantea sclerotia exudates. Nevertheless, further research is required to investigate factors that influence bacterial density and structure in this ecological niche.
Table 2 Bacterial density (Log10UFC g-1) in C. gigantea sclerotia by sampling location.
| Localidad | Número de colonias | Log10UFC g-1 de esclerocio |
|---|---|---|
| Almoloya de Juárez | z204.1 | 2.309 |
| Atlacomulco | 249.9 | 2.397 |
| Calimaya | 188.8 | 2.276 |
| Mina México | 200.4 | 2.301 |
| Toluca | 133.6 | 2.125 |
| Villa Victoria | 105.6 | 2.023 |
Z Average number of colonies isolated from 10 C. gigantea sclerotia per location sampled by direct plate count on R2A medium with three replicates
In vitro antagonism against Fusarium spp.
Out of 129 morphologically different bacterial strains isolated from sclerotia, 22 (17%) were identified by their in vitro antagonism (inhibition halo of fungal growth ≥ 5 mm) against one or more species of Fusarium (Table 3). Among these, 13 bacterial strains (59%) were isolated from the Atlacomulco location, and three strains (13%) from each of the Calimaya, Mina, and Toluca locations. Among the antagonists (n=22), 10 (45%) strains were antagonistic to F. verticillioides, 14 (63%) to F. subglutinans, and 17 (81%) to F. graminearum. Among the antagonistic bacteria, the strains Bacillus subtilis (BA1), Pseudomonas syringae (BA2), and Bacillus amyloliquefaciens (BA18) stood out, which were antagonistic against the three evaluated species of Fusarium. These three strains originated from the Atlacomulco location (Table 3).
Table 3 Molecular identification, origin, antagonism against Fusarium spp. and metabolite production of 22 antagonistic strains isolated from sclerotia of Claviceps gigantea.
| Antagonismo in vitro | Producción de metabolitos in vitro | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID cepa | Localidad | Identificación por secuenciación parcial del gen 16S rRNA | No. acceso de secuencias tipo de la especie (NCBI) | % de identidad | zFg | Fs | Fv | AIA | LIP | PRO | SID | SFM |
| BA1 | Atlacomulco | Bacillus subtilis | KF021537.1 | 99.39 | + | + | + | + | + | + | + | + |
| BA2 | Atlacomulco | Pseudomonas syringae | NR_043716.1 | 99.28 | + | + | + | + | + | + | + | + |
| BA3 | Atlacomulco | Delftia lacustris | KF054933.1 | 99.46 | + | - | + | - | + | + | + | + |
| BA4 | Mina | Stenotrophomonas sp. | AM913974.1 | 99.40 | + | - | + | - | + | + | + | + |
| BA5 | Calimaya | Delftia acidovorans | EF564190.1 | 99.38 | + | - | + | - | + | + | + | + |
| BA6 | Calimaya | Sphingobacterium sp. | KF777439.1 | 99.41 | + | - | - | - | + | + | + | + |
| BA7 | Atlacomulco | Pseudomonas geniculata | JX042457.1 | 99.44 | + | - | - | - | + | + | + | + |
| BA8 | Mina | Micromonospora sp. | KY015111.1 | 99.44 | - | + | - | - | + | + | + | + |
| BA9 | Mina | Stenotrophomonas maltophilia | FJ859699.1 | 99.37 | - | + | - | - | + | + | + | + |
| BA10 | Calimaya | Staphylococcus aureus | LN929738.1 | 99.49 | + | + | - | - | + | + | + | + |
| BA11 | Atlacomulco | Pseudomonas putida | KC582298.1 | 99.05 | + | + | - | + | + | + | + | + |
| BA12 | Toluca | Bacillus sp. | HM032893.1 | 99.48 | + | + | - | + | + | + | + | + |
| BA13 | Atlacomulco | Pseudomonas fluorescens | GU198115.1 | 99.47 | + | + | - | + | + | + | + | + |
| BA14 | Atlacomulco | Bacillus subtilis | KF527828.1 | 99.35 | + | + | - | - | + | + | + | + |
| BA15 | Atlacomulco | Bacillus amyloliquefaciens | KC494392.1 | 99.02 | + | + | - | + | + | + | + | + |
| BA16 | Atlacomulco | Pseudomonas putida | KC582298.1 | 99.16 | + | + | - | + | + | + | + | + |
| BA17 | Atlacomulco | Pseudomonas fluorescens | GU198113.1 | 99.40 | - | + | - | + | + | + | + | + |
| BA18 | Atlacomulco | Bacillus amyloliquefaciens | MH781489.1 | 99.02 | + | + | + | + | + | + | + | + |
| BA19 | Atlacomulco | Pseudomonas sp. | DQ991143.2 | 99.05 | - | + | + | - | + | + | + | + |
| BA20 | Toluca | Bacillus amyloliquefaciens | MF765339.1 | 99.48 | + | - | + | + | + | + | + | + |
| BA21 | Atlacomulco | Pseudomonas putida | JX120503.1 | 99.15 | + | - | + | + | + | + | + | + |
| BA22 | Toluca | Bacillus sp. | MF510169.1 | 99.48 | - | - | + | + | + | + | + | + |
z Fg= Fusarium graminearum; Fs=Fusarium subglutinans; Fv=Fusarium verticillioides; IAA=indole-3-acetic acid production; LIP= lipolytic activity; PRO= proteolytic activity; SID= siderophore production; SFM= mineral phosphate solubilization.
Molecular identification of antagonists
The identification of the 22 antagonist strains was made possible through partial amplification of the 16S rRNA gene, which showed a similarity range of 99.02% to 99.49% when aligned against the NCBI gene bank (refer to Table 3). These strains belonged to various genera, including Pseudomonas (36.3%), Bacillus (31.8%), Delftia (9.09%), Stenotrophomonas (9.09%), Micromonospora (4.5%), Sphingobacterium (4.5%), and Staphylococcus (4.5%). Notably, P. putida (37.5%), P. fluorescens (25%), P. geniculata, and P. syringae (12.5%) were identified among the Pseudomonas species, while B. amyloliquefaciens (57.1%) and B. subtilis (28.5%) were the most common among the Bacillus species. It is worth mentioning that the majority of the antagonists (n=13) were isolated from Atlacomulco (59%), while the remaining strains (n=3) (14.3%) were obtained in equal proportions from Calimaya, Mina, and Toluca, respectively (refer to Table 2). Taxonomically, these 22 strains were distributed across four phyla and six orders, with the Firmicutes (Gram-positive) and Proteobacteria (Gram-negative) phyla, as well as the Bacillales and Pseudomonadales orders, having a higher abundance of Bacillus and Pseudomonas genera. Conversely, the Micromonospora, Sphingobacterium, and Staphylococcus genera were less abundant, with the Firmicutes (Gram-positive) and Bacteroidetes (Gram-negative) phyla, as well as the Actinomycetales, Sphingobacteriales, and Bacillales orders, having the least number of strains (refer to Table 4).
Table 4 Taxonomic distribution of 22 bacterial strains antagonistic in vitro against Fusarium spp. isolated from sclerotia of C. gigantea.
| Phylum | Clase | Orden | Familia | Género | Frecuencia % |
|---|---|---|---|---|---|
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | 36.3 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus | 31.8 |
| Proteobacteria | Beta Proteobacteria | Burkholderiales | Comamonadaceae | Delftia | 9.1 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Stenotrophomonas | 9.1 |
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Sphingobacteriaceae | Sphingobacterium | 4.5 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micromonosporaceae | Micromonospora | 4.5 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | 4.5 |
Qualitative in vitro production of metabolites
The 22 antagonists showed in vitro lipolytic and proteolytic activity, produced siderophores, and solubilized mineral phosphate; however, only 12 (55%) produced indole-3-acetic acid. Only the strains Bacillus subtilis (BA1), Pseudomonas syringae (BA2), and Bacillus amyloliquefaciens (BA18) were antagonistic against the three species of Fusarium and produced all the evaluated metabolites (Figure 2, Table 3).
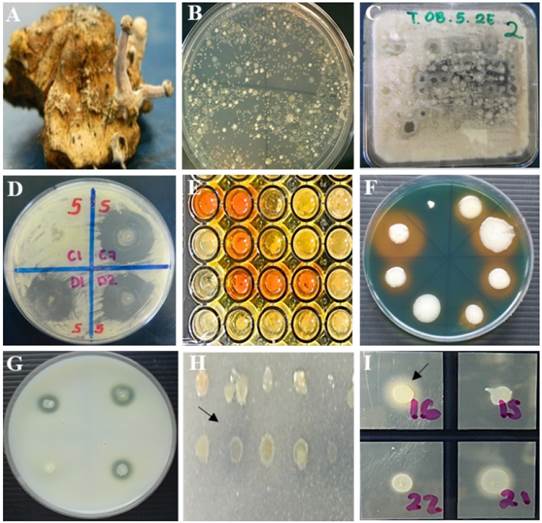
Figure 2 In vitro antagonism. A) Sclerotia of C. gigantea, B) bacterial growth from 1 g of sclerotia on R2A medium, C) in vitro antagonism of bacteria in Waksman agar culture medium inoculated with a multipoint inoculator (Boekel®, 96-point microplate replicator 96 points) against Fusarium spp. D) selection of antagonistic bacterial strains showing inhibition zones ≥5 mm of fungal growth of Fusarium spp. In vitro production of metabolites by antagonistic strains. E) production of indole-3-acetic acid, F) production of siderophores, G) proteolytic activity, H) mineral phosphate solubilization, I) lipolytic activity. Strains that showed reddish coloration of the medium, yellow, clear and opaque halo around the colony, respectively, were considered positive.
Discussion
In this study, we estimated bacterial density in C. gigantea sclerotia and evaluated their in vitro antagonism against three Fusarium species causing ear rot in maize. Our results showed varying bacterial densities associated with C. gigantea sclerotia across the different sampled locations. The highest bacterial density was recorded in the lots of Atlacomulco (2.397), while the lowest was observed in Villa Victoria (2.023) Log10CFU g-1 of sclerotium. This variation may be attributed to the use of agrochemical products and/or residues as part of the agronomic management of the crop, which may have affected the diversity and density of microorganisms associated with maize cultivation (FAO, 2010). Thus, there is a need for further research to examine the impact of agrochemical residues used in the sampled sites on the structure and diversity of bacterial communities in sclerotia. Additionally, it has also been shown that bacterial density is closely related to the sources of carbon contained in sclerotia exudates (Coley-Smith and Dickenson, 1971). Hence, future research should focus on identifying the sources of carbon present in C. gigantea sclerotia exudates and their quantity. Previous studies have shown that bacterial populations are associated with sclerotia. Coley-Smith and Dickenson (1971) found that the Sclerotium cepivorum fungus sclerotia favor the growth of specific bacteria due to the exudates containing carbon sources such as ethanol, trehalose, glucose, and mannitol, and that this microenvironment constitutes a specific ecological niche. Similarly, Gilbert and Linderman (1971) reported qualitative changes and increased microbial activity in soil near Sclerotium rolfsii sclerotia attributed to the exudates in the sclerotia of this pathogen. These authors coined the term “mycosphere” to describe the portion of soil influenced by sclerotia, which harbors bacterial populations with a greater representation of antagonistic species against S. rolfsii than those obtained from other ecological niches.
In this study, 129 bacterial strains were isolated from sclerotia in various sampled locations. Of these, 22 (17%) showed in vitro antagonism against one or more Fusarium species that cause maize ear rot, indicated by a fungal growth inhibition halo of ≥ 5 mm. Through partial sequencing of the 16S rRNA gene, it was possible to identify the 22 antagonistic bacterial strains as belonging to the genera Bacillus, Delftia, Micromonospora, Pseudomonas, Sphingobacterium, Staphylococcus, and Stenotrophomonas.
The most abundant antagonists in C. gigantea sclerotia belonged to the genera Pseudomonas (Pseudomonadaceae) (36.3%) and Bacillus (Bacillaceae) (31.8%). In both genera, their mechanisms for phytopathogen biocontrol mediated by antibiosis, competition for nutrients and space, promotion of growth, and induction of resistance in plants have been elucidated (Cui et al., 2019; Dimkić et al., 2022; Fira et al., 2018; Guzmán-Guzmán and Santoyo, 2022; Luo et al., 2022; Singh et al., 2022).
The findings of this investigation align with those of previous studies, which have highlighted the prevalence of both Pseudomonas (Pseudomonadaceae) (36.3%) and Bacillus (Bacillaceae) (31.8%) genera colonizing Rhizoctonia solani sclerotia (Zachow et al., 2011) and Sclerotium cepivorum (Backhouse and Stewart, 1989; Utkhede and Rahe, 1980), demonstrating their strong antagonism against these pathogens. Stenetrophomonas genus was also identified in this research. Wong and Hughes (1986) have shown that Bacillus species constitute 80% of antagonists isolated from soil and S. cepivorum sclerotia samples. This genus includes a group of Gram-positive bacteria with high phenotypic and genetic heterogeneity, considered among the most common colonizers in various ecological niches (Abriouel et al., 2011). Although most of the identified strains in this study were in vitro antagonistic against one or two Fusarium species, Bacillus amyloliquefaciens (BA18), B. subtilis (BA1), and Pseudomonas syringae (BA2) strains were antagonistic against all three evaluated Fusarium species (F. graminearum, F. subglutinas, and F. verticillioides) causing maize ear rot.
In other studies, B. amyloliquefaciens has been reported as an antagonist with high potential for the biocontrol of other Fusarium species in spinach (Spinacia oleracea) (Zhao et al., 2014), banana (Musa sp.) (Tian et al., 2021), tomato (Solanum lycopersicum) (Elanchezhiyan et al., 2018; Proca et al., 2020), and wheat (Triticum sp.) (Ursan et al., 2019). Some strains of B. amyloliquefaciens have been reported as antagonists and efficient biocontrol agents against F. graminearum (de Ángel et al., 2021; Gu et al., 2017; Liu et al., 2019) and F. verticillioides (Xu et al., 2021).
The prevalence and antagonism of B. subtilis isolated from sclerotia of C. gigantea coincides with other studies. Utkhede and Rahe (1980) showed that there is a greater prevalence of B. subtilis in S. cepivorum sclerotia collected from various parts of the world and that most of these strains significantly protected onion (Allium cepa) from white rot caused by this pathogen when inoculated into seed at the time of planting. These authors attributed the prevalence of B. subtilis to specific carbohydrates contained in the exudates of S. cepivorum sclerotia. Also, strains of B. subtilis have shown antagonism and efficient biocontrol against F. graminearum and F. verticillioides in wheat and maize, respectively (Cavaglieri et al., 2005; Guimarães et al., 2021; Wang et al., 2020; Yu et al., 2021).
The genus Pseudomonas includes Gram-negative species which have been widely studied as an alternative in the biological control of phytopathogens and promotion of plant growth (Dimkić et al., 2022; Guzmán-Guzmán and Santoyo, 2022; Singh et al., 2022). Pseudomonas syringae, identified in this study as an antagonist against the three species of Fusarium, belongs to the P. fluorescens species complex and has been described as an important biocontrol agent along with P. aeruginosa, P. aureofaciens, P. chlororaphis, P. fluorescens, and P. putida (Panpatte et al., 2016). Pseudomonas syringae is a species complex that includes phytopathogenic strains on a wide range of host plants (Baltrus et al., 2017). However, non-phytopathogenic strains of P. syringae have been identified whose genome harbors an extensive group of genes related to the biocontrol of phytopathogens, promotion of growth, and induction of resistance in plants (Passera et al., 2019). Yu et al. (2017) showed that the Pseudomonas syringae BAF.1 strain completely inhibited the germination of conidia and affected the structure of the mycelium of Fusarium oxysporum, proposing it as a promising biocontrol agent against this pathogen.
In this research, Bacillus subtilis (BA1), Pseudomonas syringae (BA2), and Bacillus amyloliquefaciens (BA18) strains were antagonistic against all three Fusarium species and produced in vitro all evaluated metabolites (indole-3-acetic acid production, lipolytic and proteolytic enzymes, siderophores, and mineral phosphate solubilization) (Table 3). The production of these metabolites has been shown to play an important role in the biocontrol of phytopathogenic fungi and promotion of plant growth (Sagar et al., 2022). Proteolytic and lipolytic enzymes produced by Bacillus and Pseudomonas species cause cellular lysis of fungi; siderophores are antimicrobial compounds that facilitate iron mobilization and solubilization of nutrients not available to plants (Admassie et al., 2022). The production of plant growth regulators such as indole-3-acetic acid stimulates the development of the plant root system and induces resistance to pathogens; while mineral solubilization such as phosphorus promotes plant development, induces resistance to pathogens, and improves water and nutrient uptake (Mahapatra et al., 2022; Sagar et al., 2022).
The ability of Bacillus and Pseudomonas species to produce a wide range of secondary metabolites encoded by various gene clusters has been suggested to result in disease suppression, growth promotion, and induction of resistance in plants (Andrić et al., 2020; Dimkić et al., 2022; Luo et al., 2022). However, it is important to note that the proportion of genes involved in the synthesis of antimicrobial compounds and other bioactive secondary metabolites varies depending on the species and strain (Devi et al., 2019). Therefore, prior to registering their 16S rRNA sequences in the NCBI GenBank, it is recommended to further investigate the genome characteristics of the three antagonistic strains identified in this research. Previous studies have shown that inoculation of Bacillus and Pseudomonas spp. strains promotes growth and induces resistance in maize plants (Egamberdiyeva et al., 2007).
Species of the genus Bacillus are promising biological control agents due to their genetic characteristics, high heat and desiccation resistance through the formation of endospores (Luo et al., 2022). These strains are considered to be safe for human health, hence there are no restrictions on their use as biocontrol agents according to the United States Environmental Protection Agency (Dimkić et al., 2022; Hou et al., 2006). Additionally, commercial formulations of P. syringae are approved for the management of Fusarium spp. in post-harvest in the USA and Canada (Al-Mughrabi et al., 2013). Therefore, the Bacillus subtilis (BA1), Pseudomonas syringae (BA2), and B. amyloliquefaciens (BA18) strains identified in this research, isolated from sclerotia of C. gigantea that were antagonistic in vitro against three Fusarium species causing corn ear rot, and multifunctional in the production of secondary metabolites, represent an important biotechnological resource for future investigations as biocontrol agents against this pathogen in corn cultivation in Mexico.
Conclusions
The findings of this study suggest that the sclerotia of C. gigantea contain varying densities of bacterial populations, with those from the Atlacomulco locality exhibiting the highest density. Among these bacterial populations are strains that exhibit antagonism against at least one species of Fusarium, known to cause corn ear rot and produce metabolites that may promote plant growth. Through sequencing of the 16S rRNA gene, Bacillus and Pseudomonas genera were identified as the most abundant antagonistic bacteria in C. gigantea sclerotia. In vitro experiments demonstrated that Bacillus subtilis (BA1), Pseudomonas syringae (BA2), and Bacillus amyloliquefaciens (BA18) strains were effective antagonists against all three species of Fusarium evaluated, with Fusarium graminaerum being the most susceptible. These results suggest that these strains may represent a viable option for biocontrol of Fusarium species in maize cultivation in Mexico.











 texto en
texto en 



