Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias pecuarias
versión On-line ISSN 2448-6698versión impresa ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.14 no.1 Mérida ene./mar. 2023 Epub 24-Mar-2023
https://doi.org/10.22319/rmcp.v14i1.6014
Articles
Molecular detection of a fragment of bluetongue virus in sheep from different regions of Mexico
a Instituto Nacional de Investigaciones Forestales Agrícolas y Pecuarias (INIFAP). Centro Nacional de Recursos Genéticos. México.
b INIFAP. Centro Nacional de Investigación Disciplinaria en Salud Animal e Inocuidad (CENID-SAI), Campus Ciudad de México. Carretera México-Toluca Km 15.5, Colonia Palo Alto. 05110. Alcaldía Cuajimalpa de Morelos. Ciudad de México. México.
c Universidad Nacional Autónoma de México, FMVZ. México.
Bluetongue disease (BTD) affects various species of wild and domestic ruminants. In Mexico, the disease, caused by the bluetongue virus (BTV) is still regarded as exotic, despite the fact that antibodies have been detected on several occasions. The objective was to establish molecular techniques using a synthetic gene, including the genes NS1 and NS3 as positive controls for the diagnosis of BTV in samples of sheep from different regions of the country. A total of 320 total whole blood samples were obtained from sheep. The samples obtained were evaluated by end-point RT-PCR and real-time RT-PCR, the conditions having been established by the work group. Twelve sheep samples were found to be positive for the detection of NS1; these samples were sequenced, and a fragment of 101 base pairs was obtained. Upon alignment, were obtained identities with sequences reported in GenBank with NS1 fragments ranging from 89 % (p= 1e-12) to 98 % (p= 4e-13), corresponding to serotypes 10, 11 and 12. From these samples, two positive sheep samples were obtained using real-time PCR (RT-PCR): one from Chiapas (Chiapas breed), and the other, from Tamaulipas (Suffolk breed). The results of the RT-PCR were corroborated by CPA-SENASICA. This work provides evidence, for the first time in Mexico, of the importance of using a synthetic gene as a positive control to perform BSL-2 detection in official laboratories, which in a health emergency is of utmost importance.
Key words Bluetongue disease; Bluetongue virus; Diagnosis; NS1 and NS3 genes; Sheep; Synthetic gene
La enfermedad de la lengua azul (LA) afecta diferentes especies de rumiantes silvestres y domésticos. En México, la enfermedad producida por el virus de la lengua azul (VLA), aún es reconocida como exótica, a pesar de que, en diferentes ocasiones se han detectado anticuerpos. El objetivo fue establecer técnicas moleculares usando un gen sintético que incluye los genes NS1 y NS3 como control positivo para establecer el diagnóstico del VLA en muestras de ovinos de diferentes regiones del país, mediante técnicas moleculares. Se obtuvieron 320 muestras totales de sangre completa de ovinos. Las muestras obtenidas se evaluaron mediante RT-PCR punto final y RT-PCR en tiempo real estableciendo las condiciones por el grupo de trabajo. Se encontraron 12 muestras positivas de ovinos a la detección de NS1; estas muestras se secuenciaron obteniendo un fragmento de 101 pares de bases. Al realizar el alineamiento se obtuvieron identidades con secuencias reportadas en el GenBank con fragmentos de NS1 desde 89 % (p= 1e-12) a 98 % (p= 4e-13), correspondientes a los serotipos 10, 11 y 12. De estas muestras, se obtuvieron dos muestras positivas de ovinos mediante el PCR tiempo real (PCR-tr), uno proveniente de Chiapas (raza Chiapas) y el otro de Tamaulipas (raza Suffolk). Los resultados de la PCRtr fueron corroborados por la CPA-SENASICA. Este trabajo, aporta por primera vez en México, la importancia de usar un gen sintético como control positivo, para realizar la detección en laboratorios oficiales BSL-2, lo cual en una emergencia sanitaria es de suma importancia.
Palabras clave Lengua azul; Ovinos; Diagnóstico; Gen sintético; Genes NS1 y NS3
Introduction
Bluetongue virus (BTV) belongs to the genus Orbivirus and the family Reoviridae, and causes bluetongue disease (BTD) affecting both domestic and wild ruminants1. The virus has a negative-sense double-stranded RNA (dsRNA) genome consisting of 10 segments2. It is a non-enveloped virus with an icosahedral capsid, with a diameter of approximately 90 nm. The genome codes for the structural proteins that make up the external and internal capsid or core (VP1 - VP7), and the four non-structural proteins, called non-structural (NS) that are involved in the replication, maturation, or exit of the virion from infected cells). Nonstructural genes are highly conserved across the genus3,4. The NS1 gene encodes for a protein of the same name, which is expressed in the largest quantity during BTV replication and is the most abundant cytoplasmic protein. On the other hand, the NS3 gene encodes for the NS3 protein that acts as a viroporin, which is related to cell lysis3,4. Because of the above, the two genes have been used as targets in screening assays for the identification of BTV5.
The virus is transmitted by the bite of mosquitoes of the genus Culicoides spp; therefore, the occurrence of the disease is associated with the spread of this vector, although other vectors such as ticks have been reported6; it is known that the virus can remain viable throughout the life of the vector.
Currently, 28 different BTV serotypes have been described worldwide7, and the virus is distributed in practically all countries where cattle and sheep are raised. Bluetongue disease (BTD) can occur both subclinically and clinically, especially in sheep, since in cattle it is mostly asymptomatic. In countries where the disease is endemic, it causes severe economic losses to producers8. The name "blue tongue" was given to this disease by Africans who observed cyanosis on the tongue of some animals; however, this sign is not observed in all infected animals, as the signs vary between species and depend on the strain. Lesions such as hyperemia and edema of the lips and face, oral erosions and ulcers, and the typical cyanosis of the tongue are due to infection of the endothelial cells that allow increased cell permeability9.
The World Animal Health Organization (OIE)10 classifies bluetongue disease as a notifiable disease; therefore, timely diagnosis is important. The degree of severity of the disease depends on the serotype, the virus strain, and the species, age and immune status of the animal, with sheep and white-tailed deer being the most affected11; in sheep, the incubation period of BTV is six to eight days; on the other hand, cattle rarely show clinical signs, but maintain a prolonged viraemia12. Deer can also be infected by a closely related orbivirus responsible for epizootic hemorrhagic disease13. BTD is not contagious and is only transmitted by Culicoides insects; its distribution is therefore associated with the prevalence of the vector. Up to five serotypes have been identified in North America; however, seven serotypes have been reported only in the United States14. The occurrence of the virus in the Americas is mainly associated with the presence of two vector species, C. sonorensis and C. insignis15. In Mexico, although the disease is considered exotic, in the 1980s, positive serology to the virus was reported in both sheep and cattle in different regions of the country16,17. On the other hand, in 2015, the detection of a viral genome fragment in three Culicoides species (C. variipennis, C. sonorensis, and C. occidentalis) was published18. The VP2 gene is used to define the 28 BTV serotypes described so far7.
Finally, in February 2021, the Secretary of Agricultural Development, Fisheries and Aquaculture (SEDPA) of Oaxaca in the Municipality of San Pedro Mixtepec notified the CPA of oral lesions in sheep. The CPA detected two bluetongue virus-positive samples using the RT-PCR technique and the notification was made in the CPA's AVISE Newsletter19.
BTD is notifiable to the OIE, mainly because new outbreaks lead to movement and trade restrictions, resulting in severe economic losses. However, active surveillance is implemented worldwide to detect BTV infection through different tests such as virus isolation or other screening or serological tests11. The detection method par excellence is the isolation of the virus in permissive cell cultures, for subsequent genetic analysis of the virus to determine the serotype present in the sample of the affected animal. In virus endemic areas, vector control is recommended to prevent the spread of the virus, in addition to vaccination programs. Live attenuated vaccines have been used in the United States and Europe. The objective of this work was to establish molecular techniques using a synthetic gene as a positive control that included the NS1 and NS3 genes, in order to subsequently evaluate sheep samples from different regions of the country.
Material and methods
Samples
A convenience sampling of apparently healthy sheep was carried out, obtaining 3 ml of whole blood with anticoagulant (heparin) from 320 individuals from five states of the country (Chiapas, Coahuila, Estado de México, Morelos, and Tamaulipas). Samples were obtained during the summer of 2016 to 2018. Sampling was performed on males and breeding females between one and five years of age. Table 1 describes the total number of samples analyzed.
Table 1 Blood samples obtained from sheep in five Mexican states analyzed for molecular detection of bluetongue virus by RT-PCR
| State | Species | Sex | Breed | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Chiapas | Ovinos | H | Criollo | 20 | ||||
| M | Criollo | 5 | ||||||
| S/D | S/D | 66 | ||||||
| Coahuila | Ovinos | H | Cruza | 70 | ||||
| Dorper | 37 | |||||||
| Suffolk | 13 | |||||||
| Ovinos | H | Blackbelly | 1 | |||||
| Criollo | 29 | |||||||
| Pelibuey | 1 | |||||||
| Morelos | Ovinos | S/D | S/D | 62 | ||||
| Tamaulipas | Ovinos | H | Pelibuey | 8 | ||||
| Dorset | 4 | |||||||
| Suffolk | 4 | |||||||
| Total | 320 | |||||||
F= female; M= male; N/D= no data.
Synthetic gene
Since BTD is considered exotic in Mexico, a synthetic gene was designed to be used as a positive control in order to avoid using the inactivated virus or its genetic material in a BSL2 laboratory. For this purpose, two fragments of the viral genome were inserted into the pUC57 vector (GeneScript, USA) -one corresponding to the NS1 gene (354 bp), and the other, corresponding to the NS3 gene (300 bp)-, based on the BTV-11 sequences reported in GenBank (KF986511 and KM580467, respectively). For use as a positive control in the molecular assays, the plasmid concentration was set to 100 ng/µl.
Genetic material extraction
Viral RNA was extracted from 250 l of the blood sample using the Trizol LS® reagent (Ambion, USA), following the manufacturer's instructions with some modifications to the protocol. The RNA obtained was stored at -70 °C until use.
Molecular assays
Constitutive gene. In order to verify the quality of the RNA thus obtained, a fragment of the constitutive GAPDH gene was amplified by RT-PCR using the primers and conditions reported by González-Arto M, et al20. Complementary DNA synthesized from viral RNA with the M-MLV Reverse Transcriptase kit was used as a template (Invitrogen, USA).
Detection of a fragment of gene NS3. The RNA extracted from the samples served as a template for the detection of a fragment of the NS3 gene of orbiviruses using a pair of primers and the probe recommended in the OIE Manual10. RT-PCR was carried out according to a one-step amplification protocol established in our laboratory, utilizing the iTaq Universal Probe One-Step Kit (Bio-Rad, USA).
Detection of a fragment of gene NS1. In order to corroborate the presence of the BTV genome in real-time PCR positive samples, a protocol was established for the detection of a fragment of the NS1 gene using primers described in the OIE Manual10. The iProof HF Master Mix Kit was utilized for this purpose (Bio-Rad, USA). The products for the amplification of positives to this protocol were purified in agarose gels and sequenced according to the Sanger method at the IBT-UNAM Synthesis and Sequencing Unit.
Sequencing. Sequencing results were analyzed with NCBI's BLAST tool. The obtained sequences were compared with 29 sequences reported in the Gene Bank of bluetongue virus and with sequence AM745001.1 for the epizootic hemorrhagic fever virus as an outgroup. The alignment was performed using the “Multiple alignment program for amino acid or nucleotide sequences” (MAFFT version 7, AIST). A phylogenetic analysis was performed using Bayesian methods (Markov Chain Monte Carlo) and the alignment was carried out with Mesquite in MrBayes software (Open source).
Results
As an assay to evaluate the quality of the genetic material, the amplification of a fragment of approximately 400 bp of the ovine GAPDH gene was carried out as previously above. All samples used for detection of a viral genomic fragment were positive for GAPDH amplification by RT-PCR, which indicates that the genetic material was intact and in good condition for use in RT-PCR assays (Figure 1).
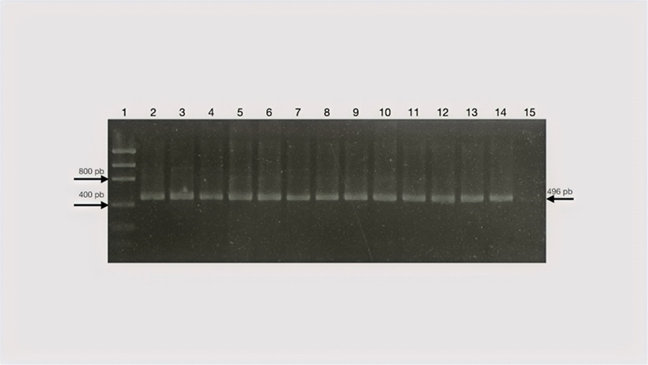
Lane 1, 50bp fragment size marker (Low Mass ladder); Lanes 2-7, sheep samples. Amplification products were run on a 1.5% agarose gel.
Figure 1 RT-PCR for amplification of the sheep’s constitutive GAPDH gene
As for the assay for the detection of a fragment of the NS1 gene by RT-PCR endpoint, the identification of this gene was obtained in 12 samples of sheep blood from the states of Chiapas, Coahuila, and Tamaulipas. In order to harmonize the methods, it was decided to use primers suggested by the OIE, as described above. The analysis of the sequences obtained showed in the alignment an identity with sequences reported in GenBank with the NS1 fragment from 89 % (p= 1e-12) to 98 % (p= 4e-13), corresponding to serotypes 10, 11 and 12.
Figure 2 shows the results of the phylogenetic inference, depicting the clustering of the samples primarily with serotypes 10 and 11. The positive results were corroborated by real-time PCR in two of the samples, one from Chiapas and the other from Tamaulipas; as mentioned above, RT-PCR uses the gene NS3.
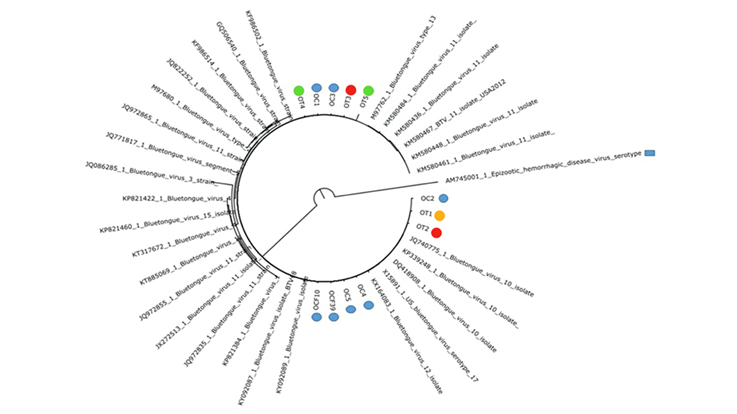
The dendogram was obtained using the alignment of 100 bp of the NS1 gene from the sequences of the positive samples in this study and 30 sequences obtained from GenBank belonging to serogroups 10, 11, and 12. Sheep samples positive by end-point RT-PCR are identified as follows: OT= Tamaulipas sheep: OT1, OT2, OT3, OT4, OT5. OC= Chiapas sheep: OC1, OC2, OC3, OCF10, OCF39, OC5, OC4. The breeds are indicated by color: Creole , Pelibuey
, Pelibuey , Suffolk
, Suffolk  , and Dorper
, and Dorper  . The samples were taken between 2016 and 2017. The sequence of the epizootic hemorrhagic fever virus (AM745001) was used as outgroup, indicated by
. The samples were taken between 2016 and 2017. The sequence of the epizootic hemorrhagic fever virus (AM745001) was used as outgroup, indicated by  .
.
Figure 2 Phylogenetic inference of bluetongue virus NS1-positive sheep samples
The positive result of RT-PCR detection was corroborated by the SENASICA - CPA laboratory and notified to SIVE. In this laboratory, the detection of a fragment of gene NS3 also serves as a positive control, using viral RNA obtained from the supernatant of cell cultures infected with the virus that are subsequently inactivated (CPA personal communication). With respect to the tests performed in collaboration with the official laboratory to corroborate the results of the animals that tested positive, the synthetic control proposed in this work showed that it could be used without problem in any BSL2 laboratory in order to perform virus detection in official BSL2 laboratories.
Discussion
As mentioned above, BTD in different hosts can be subclinical, and detection of the causative agent in sheep populations can be complex21. Therefore, for this study, clinically healthy animals were considered for sampling, where the status of viraemia in animals and signs may or may not have been observed, depending on the viral load or subtypes involved. In addition, sampling was carried out in those Federal Entities of the country located in areas where the vector transmitting the virus is present in a climate suitable for its development22, from sheep that were close to cattle farms, as bovines can be a healthy carrier of the virus.
Regarding the diagnosis of the causative agent of BTD, the recommended method is by virus isolation in a cell culture or in embryonated eggs23. However, different versions of RT-PCR have been developed that can be used to detect BTV, specifically the Orbivirus serogroup, and to determine the BTV serotype. These molecular approaches are much faster than traditional virological and immunological methods, which can take up to four weeks to provide information on serogroups and serotypes. Currently, there are targeted assays mainly for VP1, NS1, NS2, VP6, and NS3 proteins. None of these proteins is related to virus serotyping, and they are strongly conserved among BTV serotypes, while some, such as NS3, have a higher degree of conservation among orbiviruses. Therefore, these assays lack the potential to classify isolates24.
In addition, each technique offers a range of virus or genome detection; for instance, Bonneau et al25 report that the RT-PCR assay is capable of detecting the genome within a period ranging from 3 to 122 days. Therefore, it is important to make the recommendation to carry out sampling and surveillance campaigns not only on ruminants but also on the potential vectors that are reported as transmitters of the virus.
BTD is considered exotic in Mexico; however, this status should be reconsidered taking into account the various notifications made since the 1980s to the present date in different regions of the country; this would allow to assess the presence of the virus in different hosts and vectors using a variety of methods. In 1981, Moorhead et al26 determined the presence of antibodies by immunoprecipitation in sheep slaughtered at the slaughterhouse, finding 8.5 % positivity in serum. Subsequently, Vilchis et al (1986)27, using immunodiffusion, demonstrated 27.4 % seropositivity in the animals sampled. Stott et al28 reported seropositivity of 6 %, 35 % and 60 % in three independent studies on cattle from different states of the country. The most recent scientific publication by Lozano-Rendón JA and his work group18 proved a 14.4 % molecular detection of the NS1 gene of BTV in Culicoides vectors in the state of Nuevo León.
As for the results presented in this research, a 3.75 % positivity rate was detected in the samples of clinically healthy sheep using the same NS1 gene as Lozano-Rendón et al15. However, this study was conducted on the vector, where the probability of demonstrating the presence of the virus is greater than in sheep, where the viraemia time is shorter. This NS1 gene, as already mentioned, is one of the most conserved among the different BTV serotypes29. The results of the detection of a fragment of the viral genome in sheep samples in this study are consistent with those reported this year by the CPA19.
On the other hand, the detection rate is similar to that described in older reports using ruminant samples. The results reported in the present work, as well as those presented by other authors, show the need to change the status of the disease, as well as to implement virus surveillance systems in both the vectors and the main hosts of the virus, whether these be domestic or wild animals.
Conclusions and implications
A synthetic positive control is presented herein as an alternative to viral RNA, which can only be utilized in the BSL3 laboratory of the country's official agencies. The use of such synthetic positive control would enlarge the network of laboratories capable of implementing the viral detection technique to determine the real status of the disease in the country.
Acknowledgments
The authors are grateful to Roberto Navarro López, MSc; to Marcela Villarreal Silva, PhD; to Mariana García Plata, MSc, and to Martín García Osorio, DVM, for their collaboration in corroborating the results in the CPA-SENASICA Laboratory. This research was financed by INIFAP project No. 12583634008 and the validated form No. 914545716.
REFERENCES
1. Qing-Long G, Wang Q, Yang XY, Li DL, Zhao B, Ge GY, et al. Seroprevalence and risk factors of the bluetongue virus in cattle in China from 1988 to 2019: A comprehensive literature review and meta-analysis. Front Vet Sci 2021;7:550381 doi: 10.3389/fvets.2020.550381. [ Links ]
2. Maclachlan NJ. Bluetongue: History, global epidemiology, and pathogenesis. Prev Vet Med 2011;102(2):107- 111. [ Links ]
3. Chacko N, Mohanty NN, Biswas SK, Chand K, Yogisharadhya R, et al. A coiled-coil motif in non-structural protein 3 (NS3) of bluetongue virus forms an oligomer. Virus Genes 2015;51(2):244-251 doi 10.1007/s11262-015-1230-9. [ Links ]
4. Roy P. Bluetongue virus proteins and particles and their role in virus entry, assembly and release. Adv Virus Res 2015;64:69-123. doi.org/10.1016/S0065-3527(05)64004-3. [ Links ]
5. Schwartz-Cornil PP, Mertens V, Contreras B, Hemati F, Pascale E, Breard et al. Bluetongue virus: virology, pathogenesis and immunity. Vet Res 2008;39(5):46. doi: 10.1051/vetres:2008023. [ Links ]
6. Sperlova A, Zendulkova D. Bluetongue: a review. Vet Med 2011;56:430-452. [ Links ]
7. Bumbarov V, Golender N, Jenckel M, Wernike K, Beer M, Khinich E, Zalesky O, Erster O. Characterization of bluetongue virus serotype 28. Transb Emer Dis 2020;67(1):171-182. doi.org/10.1111/tbed.133338. [ Links ]
8. Maclachlan NJ, Drew CP, Drew CP, Darpel KE, Worwa G. The pathology and pathogenesis of bluetongue. J Comp Pathol 2009;141:1-16. doi.org/10.1016/j.jcpa.2009.04.003. [ Links ]
9. Drew CP, Heller MC, Mayo C, Watson JL, Maclachlan NJ. Bluetongue virus infection activates bovine monocyte-derived macrophages and pulmonary artery endothelial cells. Vet Immunol Immunopathol 2010;136(3-4):292-296. doi:10.1016/j.vetimm.2010.03.006. [ Links ]
10. OIE. Enfermedades, infecciones e infestaciones de la Lista de la OIE en vigor en 2019. http://www.oie.int/es/sanidad-animal-en-el-mundo/enfermedades-de-la-lista-de-la-oie-2018/ . Consultado 16 Nov, 2019. [ Links ]
11. Rojas JM, Rodríguez-Martín D, Martín V, Sevilla N. Diagnosing bluetongue virus in domestic ruminants: current perspectives. Vet Med Res Report 2019;10:17-27. [ Links ]
12. Barratt-Boyes SM, Maclachlan NJ. Dynamics of viral spread in bluetongue virus infected calves. Vet Microbiol 1994;40(3-4):361-371. doi.org/10.1016/0378-1135(94)90123. [ Links ]
13. Falconi C, López-Olvera JR, Gortázar C. BTV infection in wild ruminants, with emphasis on red deer: a review. Vet Microbiol 2011;151(3-4)209-219. doi.org/10.1016/j.vetmic.2011.02.011. [ Links ]
14. Drolet S, Rijn P, Howerth E, Beer M, Mertens P. A review of knowledge gaps and tools for Orbivirus research. Vector-borne Zoon Dis 2015;15(6): 339-347. doi: 10.1089/vbz.2014.1701. [ Links ]
15. Gay GC. Orbiviruses: A gap analysis. Vector Borne Zoonotic Dis-2015;15(6):333-334. doi:10.1089/vbz.2015.28999.cgg. [ Links ]
16. Suzan VM, Misao O, Romero EA, Yosuke M. Prevalence of bovine herpesvlrus-1, paraenfluenza-3, bovine rotavirus, bovine viral diarrhoea, bovine adenovirus 7, bovine leukemia virus and bluetongue virus antibodies in cattle in Mexico. Jpn J Vet Res 1983;31(3-4): 125-132. [ Links ]
17. Vilchis C, Gay J, Batalla D. Determinación de anticuerpos contra el virus de lengua azul en ovinos por la técnica de inmunodifusión. Tec Pecu Méx 1986;51:116-121. [ Links ]
18. Lozano-Rendón JA, Contreras-Balderas AJ, Fernández-Salas I, Zarate-Ramos J, Avalos-Ramírez R. Molecular detection of bluetongue virus (BTV) and epizootic hemorrhagic disease virus (EHDV) in captured Culicoides spp. in the northeastern regions of Mexico. Afr J Microbiol Res 2015;9(45):2218-2224. [ Links ]
19. Boletín Informativo de la CPA. AVISE. No 9 Febrero, 2021. https://issuu.com/boletinavise/docs/boletin_avise_ed09_febrero. [ Links ]
20. Gonzalez-Arto M, Hamilton dos STR, Gallego M, Gaspar-Torrubia E, Aguilar D, Serrano-Blesa E, et al. Evidence of melatonin synthesis in the ram reproductive tract. Andrology 2016;4(1):167-171. doi: 10.1111/andr.12117. [ Links ]
21. Celma CC, Bhattacharya B, Eschbaumer M, Wernike K, Beer M, Roy P. Pathogenicity study in sheep using reverse-genetics-based reassortant bluetongue viruses Vet Microbiol 2014;174(1-2):139-47. doi: 10.1016/j.vetmic.2014.09.012. [ Links ]
22. SIAP. Producción por Estado. 2016. http://infosiap.siap.gob.mx/anpecuario_siapx_gobmx/apecnal.jsp?id=5. [ Links ]
23. McHolland LE, Mecham JO. Characterization of cell lines developed from field populations of Culicoides sonorensis (Diptera: Ceratopogonidae). J Med Entomol 2003;40(3):348-51. doi: 10.1603/0022-2585-40.3.348. [ Links ]
24. Maan S, Maan NS, Belaganahalli MN, Potgieter AC, Kumar V, Batra K, et al. Development and evaluation of Real Time RT-PCR assays for detection and typing of Bluetongue Virus. PLoS ONE 2016;11(9): e0163014. doi: 10.1371/journal.pone.0163014. [ Links ]
25. Bonneau KR, DeMaula CD, Mullens BA, Maclachlan NJ. Duration of viraemia infectious to Culicoides sonorensis in bluetongue virus-infected cattle and sheep. Amsterdam: Elsevier Scientific Publishing Co; 2002. [ Links ]
26. Moorhead JR. Estudio de la presencia de anticuerpos precipitantes contra el virus de la Lengua Azul en ovinos y bovinos sacrificados en el Rastro de Ferrería de la Ciudad de México, DF [tesis]. Facultad de Medicina Veterinaria y Zootecnia, Universidad Nacional Autónoma de México;1981. [ Links ]
27. Vilchis CM, Gay GJ, Batalla D. Determinación de anticuerpos contra el virus de lengua azul en ovinos por la técnica de inmunodifusión. Tec Pecu Méx 1986;51:116-121. [ Links ]
28. Stott JL, Blanchard-Channell M, Osburn BI, Riemann HP, Obeso RC. Serologic and virologic evidence of bluetongue virus infection in cattle and sheep in Mexico. Am J Vet Res 1989;50(3):335-340. [ Links ]
29. Mertens PP, Diprose J, Maan S, Singh KP, Attoui H, et al. Bluetongue virus replication, molecular and structural biology. Vet Italiana 2004;40(4):426-437. [ Links ]
Received: July 01, 2021; Accepted: August 31, 2022











 texto en
texto en 


