1. Introduction
The Earth’s atmosphere is an intricate and ever-changing blanket of gases that envelops the surface of our planet, surrounding not only all human beings but also terrestrial plants, animals, buildings, and man-made objects. These gases carry suspended particles, both solid and liquid, along with microorganisms, which may be attached to them or in free form (Zhai et al., 2018). All these components play a crucial role in our interactions with the environment. Biological and human activities affect them, and they in turn influence biological processes.
Numerous studies have been conducted in major cities around the world to analyze the effects of the composition and size of airborne particulate matter on human health (e.g., Davidson et al., 2005; Querol et al., 2008; Chirino et al., 2015); and in recent years, interest in the study of airborne microorganisms has also increased worldwide (Song et al., 2022; Triadó-Margarit et al., 2022; Noda et al., 2023).
Based on the PubMed database (NCBI, 2023), the number of published articles on airborne microorganisms has experienced substantial growth over the past two decades. In 2000, there were 246 articles on this topic, whereas in 2023, the figure soared to 1180 papers, representing an impressive nearly five-fold increase. This was mainly due to the implications of microorganisms on human health and the deterioration of agricultural products, buildings, crops, foodstuffs, and man-made objects.
The composition and structure of airborne microbial communities exhibit continuous spatial and temporal variations influenced by various factors, including geographical features, environmental conditions, and human activities (e.g., Li et al., 2019; Qi et al., 2020, Tignat-Perrier et al., 2020). Specific investigations are needed to understand both cross-sectional and temporal variations in airborne microbiota and their underlying causes.
Several studies on airborne microbiota have been conducted in Mexico using different research approaches. This review aims to identify, organize, and analyze them to find out how many there are, how and where they have been carried out, and what they have uncovered. The analysis is divided into three sections: (1) a bibliometric investigation of the studies, (2) an examination of the methodologies employed, and (3) an analysis of the airborne microorganisms found in different sites in Mexico. On this basis, the state of the art and gaps in knowledge were identified and discussed.
2. Material and methods
A systematic review of articles up to June 2023 was performed using the Web of Science (WS) database. The search keywords used were “Airborne microorganisms Mexico” and “Airborne microbiota Mexico”. All retrieved references were reviewed and assessed based on their title and abstract to decide whether or not to include them in this study. The following studies were excluded: (1) studies not conducted in mainland Mexico (e.g., studies conducted in the Gulf of Mexico or New Mexico), (2) studies that did not investigate airborne microorganisms, (3) non-peer-reviewed papers, and (4) studies conducted in indoor environments.
To avoid overlooking any relevant research on the topic, a second search was conducted using the same keywords in the Google Scholar (GS) database, with the same inclusion and exclusion criteria, and its results were added to the WS-derived database.
The bibliographic information found was systematized in a database where the information of each article was registered, including the year of publication, sampling method, organisms studied, identification method, state of the Mexican Republic where the study was carried out, and if applicable, the culture medium used, as well as the methods of DNA extraction and sequencing. Finally, the studies were classified into three types: (1) those employing culture techniques, when a culture medium was used to isolate microorganisms for subsequent identification; (2) those based on microscopy, where the bulk particles collected were analyzed directly under the microscope; and (3) those carried out using metagenomics, which involves the collection of environmental samples, extraction of metagenomic DNA, sequencing and in silico identification of microorganisms.
3. Results and discussion
3.1. Bibliometric analysis
In the initial WS search, a total of 91 studies were identified, 69 of which were excluded because they did not meet the established criteria, and a further six studies were excluded because they were conducted in indoor environments. So finally, by adding 19 studies found in the GS search, a total of 35 studies on outdoor air microbiota in Mexico were found. The first of these was published in 1987 and the last in June 2023 (Fig. 1).
The average number of studies on airborne microbiota conducted in Mexico is about one per year, and it should be noted that in 16 of these 37 years, no paper was published (Fig. 2). However, in the last eight years there has been a marked increase in the frequency of publications, with at least one publication per year, bringing the average to 1.75 per year. Thus, it seems that publications on this topic are becoming more frequent. The years 2014 and 2016 stand out as those with the highest number of publications, with five and three articles respectively.
As can be seen in Figure 2, from 1987 to 2013, articles published in GS were more abundant than those in the WoS; but from 2013 onwards this ratio was reversed, with WoS articles taking the lead. This change was probably because WoS did not have the extensive number of indexed journals that it has today, even though it was launched in 1964 as the Science Citation Index (SCI). The platform has undergone several updates and extensions over time, and its number of indexed journal articles has increased as the number of indexed journals has grown.
Within the period studied, articles on airborne microbiota in Mexico meeting the search criteria were published in 25 journals (Table I). The journal with the highest number of articles was Aerobiologia, with a total of six. It was followed by Atmospheric Environment, Environmental Pollution, International Journal of Biometeorology, and Water, Air, and Soil Pollution, each with two studies. These journals accounted for 40% of the airborne microbiota reports in Mexico. The remaining 60% were distributed among 20 journals, with one study published in each.
Table I Scientific journals in which articles related to airborne microbiota in Mexico were published from 1987 to jun2023. Only those meeting the criteria mentioned in the text were considered.
| Journal | Number of articles | Impact factor (WoS, 2023) |
| Aerobiologia | 6 | 2.0 |
| Atmospheric Environment | 2 | 5.0 |
| Environmental Pollution | 2 | 8.9 |
| International Journal of Biometeorology | 2 | 3.2 |
| Water Air and Soil Pollution | 2 | 2.9 |
| Applied and Environmental Microbiology | 1 | 4.4 |
| Atmósfera | 1 | 1.4 |
| Atmosphere | 1 | 2.9 |
| Environmental Research | 1 | 8.3 |
| Frontiers of Environmental Science & Engineering | 1 | 6.4 |
| Geofísica Internacional | 1 | 0.4 |
| Ingeniería | 1 | NA |
| International Journal of Environmental Health Research | 1 | 3.2 |
| International Journal of Family & Community Medicine | 1 | NA |
| International Research Journal of Biological Sciences | 1 | NA |
| Journal of Aerosol Science | 1 | 4.5 |
| Journal of Environmental Biology | 1 | NA |
| Journal of Environmental Health | 1 | 0.8 |
| Journal of Environmental Protection | 1 | NA |
| Journal of Environmental Sciences | 1 | 6.9 |
| Journal of Exposure Analysis and Environmental Epidemiology | 1 | NA |
| Journal of the Mexican Chemical Society | 1 | 1.5 |
| Microbial Ecology | 1 | 3.6 |
| Revista Latinoamericana de Microbiología | 1 | NA |
| Revista Mexicana de Micología | 1 | NA |
NA, journals that are not in the Web of Science 2023 database. WoS, 2023.
3.2 Analysis of the aims pursued
Almost all articles on airborne microbiota published so far in Mexico have been aimed at expanding the basic knowledge on this topic rather than looking for a concrete practical application; however, it is worth mentioning that most have focused on the presence of potentially pathogenic or allergenic genera.
Regarding bacterial microbiota, eight of the 14 studies conducted so far aimed to investigate the presence of potentially pathogenic bacteria in the air, as well as their possible correlations with environmental variables such as spatial distribution and seasonality. Using culture techniques, they have specifically analyzed the presence of genera such as Staphylococcus, Streptococcus, Pseudomonas, Bacillus, Escherichia, Enterobacter, Enterococci, Listeria, Klebsiella, Proteus, and Citrobacter. Another study also evaluated the antibiotic resistance of Staphylococcus isolates, which was the closest to a potential application (Alvarado et al., 2012).
Three additional works also aimed to analyze the possible relationships between seasonality and airborne bacteria, but these by using metagenomics. And finally, two more studies aimed to develop or evaluate the suitability of some methodologies, one for sampling airborne microorganisms and the other for specifically detecting Enterococcus faecalis.
As for the studies on airborne fungi, 10 (out of the 20 that have been published) aimed to analyze possible relationships between potentially allergenic fungal taxa and their seasonality or spatial distribution. They studied genera such as Cladosporium, Alternaria, Aspergillus, Curvularia, and Fusarium by culturing their propagules, filaments, or spores, or by identifying spores under the microscope. In addition to these 10 papers, eight other studies focused on characterizing airborne fungal communities and analyzing their relationships with temporal or environmental variables, independently of their allergenicity, either by culture, microscopy, or metagenomics. Other work focused on the identification of airborne taxa potentially pathogenic to avocado crops (which can be considered application-oriented) and a further study aimed at developing methodologies for spore identification by PCR.
Finally, seven studies investigated the presence of airborne non-fungal eukaryotes, such as algae, amoebae, and protozoa. Four of them took a descriptive approach to the presence of potentially pathogenic microorganisms, another investigated the allergenicity of airborne algae, and the remaining two studies focused on the identification and characterization of airborne microeukaryotes without a special emphasis on their possible pathogenicity or allergenicity.
3.3. Analysis of the methodologies used.
A wide variety of methodologies for the study of airborne microorganisms were used in the 35 articles analyzed. A summary of the most used methodologies is presented in Figure 3. In general, they can be divided into three main steps: (1) the sampling method, (2) the main methodological approach, and (3) the identification strategy of microorganisms (Fig. 3).
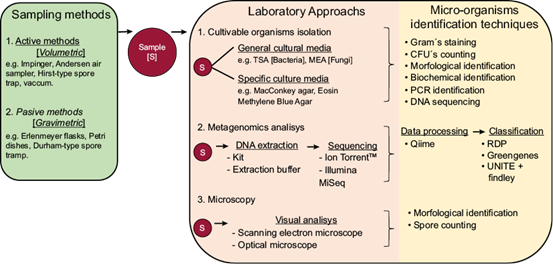
Fig. 3 Flowchart illustrating the three main methodological steps that have been used for the study of airborne microorganisms in Mexico.
3.3.1 Sampling method
Regarding the sampling method, samplers can be divided into active and passive. Active, or volumetric samplers, are devices that operate by drawing in or aspirating air through a controlled airflow. In these, the sample can be collected in different ways: (1) through multi-stage impactors which are designed to draw air in and capture particles in one or multiple stages, separating the particles according to their size for later analysis (e.g., the Andersen six-stage sampler); (2) filtered through a membrane (e.g., the high volume total suspended particulate air sampler); or (3) passed through a liquid phase (e.g., the bubble flask with culture medium or saline solution). In the analyzed publications, the airflow rate through the active samplers ranged from 10 L min-1 (for example, using a Hirst spore trap) (HTS) to 600 L min-1 (using a high throughput jet sampler), and the filtering time also had a wide range of variation (from minutes to days), so the total volume of air sampled was very different from study to study.
On the other hand, passive or gravimetric methods are devices used to capture airborne particles, contaminants, and microorganisms, without the need for active suction. These samplers are based on the natural diffusion of pollutants towards a capture surface. Examples are Erlenmeyer flasks and Petri dishes, both with general or specific culture media (solid or liquid), and Durham spore traps. Unlike active samplers, passive samplers do not require an external power source to draw in air, making them simpler and more cost-effective to use as they do not require a pump or controlled airflow.
As can be seen in Tables II and III, in both bacterial and fungal studies, active samplers have been used much more frequently than passive samplers.
Table II Main bacterial genera identified in the airborne microbiota studies conducted in Mexico (up to June 2023).
| City(s) | Sampler | Approach | Month(s) or season(s) | Main bacterial genera identified | Reference |
| Aguascalientes* | BS (A) | Culture techniques | Rainy and dry | Acinetobacter, Bordetella, Brucella, Escherichia. | Flores-Tena et al., 2007 |
| Hermosillo* | HVS (A) | Culture techniques | February, April-May, August-September | Citrobacter, Enterobacter, Enterococcus, Klebsiella, Proteus, Salmonella. | Santos-Romo et al., 2014 |
| Hermosillo*** | HVS (A) | Culture techniques | Winter, spring, and summer | Enterococcus. | Santos-Romo et al., 2019 |
| Mérida, Sisal | QT30 (A) | Culture techniques | January and July | (Actinobacteria), (Firmicutes), (Proteobacteria) | Rodríguez-Gómez et al., 2020 |
| Mexico City | AS (A) | Culture techniques | November-May and June-October | Enterobacter, Escherichia, Serratia. | Rosas et al., 1994 |
| Mexico City | AS (A) | Culture techniques | December and April | Bacillus. | Hernández-Castillo et al., 2014 |
| Mexico City | PD (P) | Metagenomics | February, March, May, June, July, September, and November | Acinetobacter, Bacillus, (Enterobacteriaceae), Erwinia, Exiguobacterium, Proteus, Pseudomonas, Staphylococcus. | García-Mena et al., 2016 |
| Mexico City | DST (P), HST (A), HJS (A) | Metagenomics | October | Acinetobacter, Calothrix, Chroococcidiopsis, Corynebacterium, Exiguobacterium, Paracoccus, Pseudomonas. | Serrano-Silva et al., 2018 |
| Mexico City | TSP (A), HST (A), HJS (A) | Metagenomics | March | Exiguobacterium, Friedmanniella, Kocuria, Methylobacterium, Microbispora, Paracoccus, Rubellimicrobium, Sphingomonas. | Calderón-Ezquerro et al., 2020 |
| Mexico City | HST (A) | Metagenomics | November-March, March-May, May-September | Corynebacterium, Kocuria, Microbispora, Paracoccus. | Calderón-Ezquerro et al., 2021 |
| Mexico City | HST (A) | Metagenomics | January-December | (Actinobacteria), (Cyanobacteria), (Firmicutes), (Proteobacteria). | Calderón-Ezquerro et al., 2022 |
| Netzahualcóyotl | PD (P) | Metagenomics | February, March, May, June, July, September, and November | Acinetobacter, Bacillus, (Enterobacteriaceae), Erwinia, Exiguobacterium, Proteus, Pseudomonas, Staphylococcus. | García-Mena et al., 2016 |
| North of Mexico** | AS (A) | Culture techniques | April, August, September | Staphylococcus | Alvarado et al., 2012 |
| Puebla* | AS (A) | Culture techniques | January-December | Citrobacter, Enterobacter, Enterococci, Escherichia, Klebsiella. | Rivera et al., 2014 |
| Tijuana | Air sampler (A) | Culture techniques | November-December | Bacillus, Enterobacter, Enterococcus, Escherichia, Klebsiella, Listeria, Pseudomonas, Staphylococcus. | Hurtado et al., 2014 |
BS= Burkard sampler, HVS= High volume sampler, QT30= Quick Take 30 Sampler, AS= Andersen sampler, PD, DST= Durham-type spore trap, HST = Hirst-type spore trap, HJS = High-throughput ‘Jet’ sampler, TSP = Total suspended particles sampler. (A)= Active sampler, (P)= Pasive sampler.
Notes: In cases where it was not possible to classify down to the genus level, the lowest taxonomic level is indicated in brackets. *The work focused on the identification of pathogenic bacteria. **The work focused on the identification of Staphylococcus aureus. *** The work focused on the identification of Enterococcus faecalis.
Table III Main fungal genera identified in the airborne microbiota studies conducted in Mexico (up to June 2023).
| City(s) | Sampler | Approach | Month(s) | Main fungal genera identified | Reference |
| Ciudad Juarez | Rotorods® (A) | Culture techniques | Jan-Sep | Alternaria, Aspergillus, Penicillium | González-Delgado et al., 2017 |
| Culiacan | MicroBio® (A) | Culture techniques | Nov-Mar | Aspergillus | Báez et al., 2014 |
| Hermosillo | BS (A) | Microscopy | Jan-May and June-Dec | Alternaria, Aspergillus, Cladosporium, Penicillium | Moreno-Sarmiento et al., 2016 |
| Hermosillo | BS (A) | Microscopy | Jan-Dec | Alternaria, Cladosporium | Ortega et al., 2019 |
| Mexico City | AS (A) | Culture techniques | Aug-Feb | Alternaria, Cladosporium, Penicillium | Rosas et al., 1990 |
| Mexico City | AS (A) | Culture techniques | Feb-May and June-Oct | Alternaria, Aureobasidium, Cladosporium, Fusarium, Penicillium | Rosas et al., 1993 |
| Mexico City | AS (A) | Culture techniques | Nov-Dec and July-Aug | Alternaria, Aspergillus, Aureobasidium Cladosporium, Eurotium, Helminthosporium | Rosas et al., 1997 |
| Mexico City | BS (A) | Culture techniques | Jan-Dec | Alternaria, Cladosporium | Calderón et al., 1997 |
| Mexico City | AS (A) | Culture techniques | Dec and Apr | Alternaria, Aspergillus, Penicillium | Hernández-Castillo et al., 2014 |
| Mexico City | AS (A) | Culture techniques | Feb, Apr and Oct | Aspergillus, Acremonium, Cladosporium, Fusarium, Penicillium | Ponce-Caballero et al., 2013 |
| Mexico City | TSP (A), HST (A), HJS (A) | Metagenomics | March | Aureobasidium, Cryptococcus, Cladosporium, Phoma | Calderón-Ezquerro et al., 2020 |
| Mexico City | HST (A) | Metagenomics | Nov-Mar, Mar-May, May-Sep | Aureobasidium, Coprinellus, Cladosporium, Phoma | Calderón-Ezquerro et al., 2021 |
| Mérida | AS (A) | Culture techniques | February | Alternaria, Acremonium, Bipolaris, Cladosporium, Fusarium | Ponce-Caballero et al., 2010 |
| Mérida | AS (A) | Culture techniques | Feb, Apr, and Oct | Acremonium, Aspergillus, Cladosporium, Fusarium, Penicillium, | Ponce-Caballero et al., 2013 |
| Mérida, Sisal | QT30 (A) | Culture techniques | Jan and July | Cladosporium, Penicillium | Rodríguez-Gómez et al., 2020 |
| Morelia | Kitasato flask/ Vacuum pump (A) | Culture techniques | Oct-Dec | Alternaria, Acremonium, Aspergillus, Cladosporium, Chrysosporium, Mucor, Monilia, Penicillium | Rivera-Tafolla et al., 2022 |
| Monterrey | AirTest (A) | Culture techniques | Feb-Jan | Aspergillus, Cladosporium, Fusarium, Penicillium | Fernández-García et al., 2023 |
| North Mexico | AS (A) | Culture techniques | Apr, Aug and Nov | Alternaria, Aspergillus, Bipolaris, Cladosporium, Rhizopous | Alvarado et al., 2012 |
| Ocuituco | PD (P) | Culture techniques | Mar-Nov | Alternaria, Capnodium, Colletotrichum, Epicoccum, Fusarium, Nigrospora, Penicillium, Ulucladium,. | Valle-Aguirre et al., 2016 |
| Toluca | Pluv (P) | Microscopy | May-Sep | Spores were not taxonomically classified. | Romero-Guzmán et al., 2021 |
Abbreviations: BS= Burkard sampler, AS= Andersen samplers, TSP = Total suspended particles sampler, HST = Hirst-type spore trap, HJS = High-throughput ‘Jet’ sampler, QT30= Quick Take 30 Sampler, PD= Petri dishes, P= Pluviometer, (A)= Active sampler, (P)= Pasive sampler.
In a study by Serrano-Silva and Calderón-Esquerro (2018), three samplers were compared: (i) a Durham-type spore trap (passive), (ii) a Hirst-type spore trap (active), and (iii) a high-throughput ‘jet’ sampler (active), and each of these three samplers was found to preferentially collect certain groups of bacteria. The passive Durham-type spore trap preferentially collected cyanobacteria, the jet sampler collected mainly proteobacteria and firmicutes, and the Hirst-type spore trap collected mainly actinobacteria, proteobacteria, and firmicutes, with the latter collecting the greatest diversity of microorganisms.
Once the collection is complete, the sample may consist of a Petri dish with a culture medium, a liquid culture medium or saline solution with microorganisms in suspension, a Melinex tape with microorganisms attached, a membrane or paper filter, or even bulk particles.
3.3.2 Laboratory approach
The second key step in the study of airborne microorganisms is the laboratory approach, which refers to the main technology used in the laboratory to analyze the microorganisms contained in the samples, and depends on the purpose of the study, the microorganisms of interest, and the facilities and financial resources available. In the 35 studies reviewed, three different laboratory approaches were identified: (i) those based on culture techniques, (ii) those using microscopy, and (iii) those using metagenomic technology (Fig. 3).
In culture-based studies, the microorganisms in the sample were grown and isolated either using general culture media, such as trypticase soy agar (TSA) for bacteria, malt extract-agar for fungi, or specific culture media such as xylose lysine deoxycholate-agar for enteric pathogens, MacConkey-agar for enterobacteria, mannitol salt-agar for staphylococci, or sodium azide esculin bile-agar for enterocococci, among others.
In turn, microscopy-based studies involved the observation of a sample of bulk particles under a microscope, which can be optical or a scanning electron microscope. However, in both cases, this technique only visualizes particles and microorganisms within specific size ranges.
As for the third approach, metagenomic studies involve the extraction of total DNA from the sample by using an extraction kit or laboratory-made solutions. This DNA is then sent for either amplicon or shotgun sequencing. In the papers we analyzed using this approach, sequencing was performed by amplicons, using as target the hypervariable regions of the rRNA16S gene for bacteria (e.g., Calderón-Ezquerro et al., 2020, 2021; Rodríguez-Gómez et al., 2020), and the ITSs regions for fungi (e.g., Calderón-Ezquerro et al., 2020). The main sequencing platforms used were Ion Torrent and Illumina-MiSeq, both considered next-generation sequencing (NGS) platforms. So far, no study has used shotgun sequencing to investigate airborne microbiota in Mexico.
It is important to note that each of the different laboratory approaches has some bias, and it is not possible to directly compare results from different studies without taking this into account. For example, metagenomic studies allow the detection of a much greater richness of microorganisms but often do not allow their reliable identification down to low taxonomic levels (such as species or strain). In contrast, culture-based techniques do not allow detection of the vast majority of microorganisms present in a sample but do allow very precise classification. That is why the results presented in Tables II and III are at the genus level. It is essential to bear this in mind when comparing results from different studies. In general, it can be said that it is possible (and important) to compare which microorganisms were present in different studies, but it is not possible to do so at low taxonomic levels, and it does not make sense to compare absences of micro-organisms between studies that used different approaches. It would be desirable to develop a protocol that would allow comparison of results between studies.
3.3.3 Microorganisms identification technique
The third key step is the identification of the microorganisms. Depending on the methodological approach followed, there is a wide diversity of identification methods. In the case of studies of culturable organisms, identification can range from morphological identification using taxonomic keys and biochemical galleries, to the identification of specific organisms of interest, such as Escherichia coli, by PCR or DNA sequencing of colonies growing on a specific culture media.
In studies following a metagenomic approach, once DNA reads are available, they are analyzed using bioinformatics procedures. This usually involves cleaning, filtering and assembling the sequences to eliminate sequencing errors and chimeras, and finally, taxonomic classification is carried out by comparing them against existing databases. The databases that have been used for taxonomic classification in studies in Mexico have been the Ribosomal Database Project (RDP), Greengenes for bacteria, and UNITE + Findley for fungi.
Finally, studies using microscopy have been based on the counting of spores collected in the environmental sample, followed by morphological identification using taxonomic keys.
Despite their differences, advantages, or limitations, the insights provided by different methodological approaches can help to gradually piece together the jigsaw of airborne microbial ecology. Of the 35 studies conducted in Mexico, 27 (77%) have been carried out using culture techniques, four (11.5%) have been carried out using microscopy, and the remaining four (11.5%) have been carried out using metagenomics. Figure 4 shows a trend where studies using culture techniques dominated until 2017, and from 2018 onwards, studies with a metagenomic approach have become increasingly frequent. Although the number of published studies using metagenomics is still quite low (totaling four, all of them published in the last six years) this is expected to increase, as the information they provide is broader. However, this change is likely to be gradual, as metagenomic studies are more expensive and the number of researchers in Mexico who have mastered this methodology is still limited. Additionally, it is worth mentioning that so far almost all metagenomic studies have been done by amplicon sequencing (16S, 18S, ITS) and have reported almost exclusively bacteria and fungi. Shotgun studies are needed to identify a wider range of microorganisms in the samples.
3.4 Results of airborne microbiota research in Mexico
3.4.1 Limitations of comparing results from different studies
Differences in sampling methods, laboratory approaches, and microorganism identification techniques make it difficult to compare results between studies. First, there is an inherent bias in any sampling method that may result in certain microorganisms being over or under-represented. Second, regarding laboratory approaches, there is a gap in the spectrum of microorganisms identified using culture techniques. It has been believed for many years that only 1% of microorganisms can be cultured using conventional culture media (Torsvik and Øvreås, 2002); however, authors such as Martiny (2019) argue that there have been significant advances in cultivation techniques in recent years and currently the percentage of culturable organisms is much higher. However, virtually all researchers agree that there is a huge gap in the microorganisms that can be identified by culture-based techniques, and that this can be minimized by using metagenomic techniques, as these can identify a broader spectrum of microorganisms in a community. Third, methodologies for the identification of microorganisms also have an important bias, as different results can be produced depending on whether morphological, biochemical, or molecular techniques are used.
The most studied group of airborne microorganisms in Mexico has been the fungal kingdom, accounting for 57% of the publications, and it is the only group that has been studied in Mexico using the three approaches discussed above. The second most studied group in terms of number of publications has been bacteria, which have been studied with both culture techniques and metagenomics. Finally, studies on archaea, microalgae, amoebae, yeasts, and protozoa are limited to four publications, and these have only been carried out by analysis of culture techniques and/or microscopy (Fig. 5).
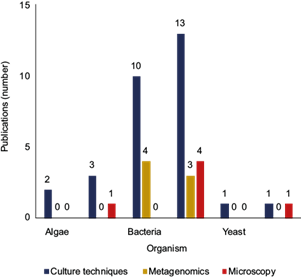
Fig. 5 Number of publications on airborne microorganisms in Mexico according to the different approaches for their study. Note: The sum of the studies does not match the total number of studies conducted (35) because some studies investigate two or more types of organisms.
Due to the great importance of both bacteria and fungi for people and ecosystems, it is to be expected that the trend in the future will be towards an equal increase in the number of studies on these two groups of microorganisms. However, this is an assumption based on what has happened internationally and not on what has happened so far in Mexico, where fungal studies have received the most attention.
3.4.2 Locations where studies have been conducted
Airborne microbiota studies in Mexico have only been conducted in 14 of the 32 states in the country: Aguascalientes, Baja California, Chihuahua, Mexico City, State of Mexico, Michoacán, Morelos, Nuevo León, Puebla, San Luis Potosí, Sinaloa, Sonora, Veracruz, and Yucatán. Furthermore, only one study has been conducted in 10 of these states, and it was performed exclusively with culture techniques (Fig. 6).
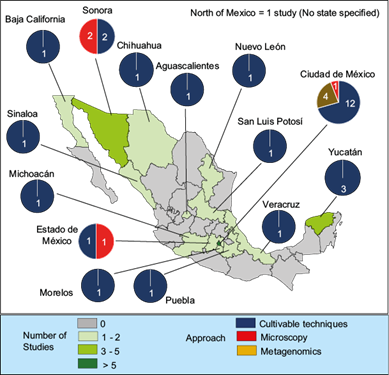
Fig. 6 Map of Mexico Map indicating the states where studies of airborne microbial communities have been reported. Note: The number of studies conducted in different states of Mexico represented on the map adds up to 37, as one study was conducted in both Mexico City and Veracruz (Rosas et al., 1989), and another study was conducted in both Mexico City and the State of Mexico (García-Mena et al., 2016).
The state where most studies on airborne microbial communities have been carried out is Mexico City, with 46% of the publications. Furthermore, it is also the only state in which studies have been carried out with a metagenomic approach, and one of only two states in which, in addition to culture techniques, studies have been carried out using microscopy (the other being Sonora).
It is worth noting that of the 35 studies conducted to date, 31 have been carried out in densely populated cities, and only four have been conducted in places uninhabited by humans (one in an avocado plantation, one in a dairy cattle production system, one in a landfill, and one in the Chihuahua desert, along the US-Mexico border). This is important because only by studying unpopulated and relatively pristine places will it be possible to have a contrast to serve as a control against which to compare the impact of cities on airborne microorganisms. So, it is to be expected that studies in other types of locations will gradually appear in the future.
3.4.3. Main bacterial genera identified
According to the findings presented in Table II, a total of 14 scientific papers have been published regarding airborne bacterial microbiota in Mexico. The majority of these studies have utilized active samplers as their primary sampling method. Nine of these investigations have employed culture techniques, while four have utilized metagenomics. It is noteworthy that approximately 40% of these studies were conducted specifically in Mexico City. Consequently, a significant knowledge gap exists regarding airborne bacterial communities in Mexico, arising from two main factors: first, the employment of culture techniques, which only allow for the identification of a small fraction of the genera present in a given sample, and second, the numerous unexplored and intriguing locations and environments within the country.
Most of these studies have focused on the identification of pathogenic bacteria. Consequently, it is not surprising that the vast majority of the genera found include species pathogenic to plants (e.g., Citrobacter, Erwinia, Pseudomonas) or to animals and humans (e.g., Acinetobacter, Bacillus, Brucella). In fact, many of the genera found belong to the order Enterobacterales (e.g., Enterobacter, Escherichia, Salmonella), which is not only associated with a wide range of clinical syndromes but is also a major causative agent of foodborne enteritis and zoonotic infections, including sporadic to pandemic outbreaks, although it is worth mentioning that is also widely dispersed in nature in many natural ecosystems (Janda and Abbot, 2021). On the one hand, this is important since all studies have been conducted in densely populated cities, and therefore imply that there is a large number of people in direct contact with airborne pathogens, but on the other hand it does not provide knowledge and understanding on the diversity, behavior, and ecological characteristics of airborne bacterial communities.
It is worth mentioning that metagenomic studies have identified a much larger number of bacterial genera than culture and microscopy studies. For example, a metagenomic study by Serrano-Silva and Calderón-Ezquerro (2018) reported 637 genera in the air of Mexico City, far from any of the studies using cultivation techniques. In addition, these methods have allowed the detection of low-abundance genera (those with < 1% relative abundance) such as Gordonia, Ruminococcus, Comamonas, Arcobacter, and Fusobacterium (Calderón-Ezquerro et al., 2020).
Most studies on airborne bacteria in Mexico have been carried out in temperate (57%) or arid (36%) regions, with tropical areas being the least studied (7%). As for the seasonality, most of them (64%) have focused on the description and general characterization of the airborne microbiota regardless of its longitudinal changes. That is, they have not compared microbial communities from the same site at different times of the year (Tables II and III). The few longitudinal studies carried out in Mexico have mainly compared the dry, wet and cold front seasons, and only a few have considered the conventional seasons (spring, summer, autumn and winter).
It is also important to note that the existing studies on airborne bacterial microbiota in Mexico have not yet generated sufficient data to discern notable trends or patterns. For instance, only very few studies have explored seasonal variations, and there is a notable dearth of information regarding altitude-based differentiations, comparisons between tropical and northern regions, or distinctions between polluted and pristine areas, among other aspects. Furthermore, limited attention has been given to investigating potential correlations between airborne bacterial communities and the physical and chemical properties of airborne particulate matter. Similarly, the influence of climatic characteristics, including humidity, temperature, and UV radiation on these communities remains significantly underexplored. Only few studies have considered the integration of environmental variables as metadata for understanding the ecology of airborne bacteria.
3.4.4. Main fungal genera identified
Fungi have been the most extensively studied group of airborne microorganisms in Mexico, with a total of 15 studies, conducted in nine of Mexico’s 32 states (Table III). As in the case of bacteria, most of the fungal studies have been conducted in state capitals or densely populated cities. In fact, only two of them fall outside this pattern, one conducted in the coastal city of Sisal, Yucatan, and the other in Ocuituco, Morelos. The latter was carried out with the aim of identifying avocado pathogens, which highlights the importance of studying airborne fungi also for their role as pathogens of economically important crops.
As in the case of bacteria, active samplers were the most commonly used to study airborne fungi, and culture techniques were the most commonly used laboratory approach (65% of studies), followed by microscopy (two studies) and metagenomic (two studies). It is interesting to note that, in addition to the studies included in this review where airborne microorganisms were collected directly from the air, fungi have been also collected indirectly from the atmosphere via spores in rainwater, as in the study by Rosas et al. (1986).
Regarding identification, the most common method was the use of taxonomic keys. It should be noted that metagenomic approaches to the study of airborne fungi have only been used in Mexico City, indicating that in most parts of the country there is still much to be discovered about the composition of airborne fungal communities and their ecology.
The genera Cladosporium and Alternaria stand out by far as the most commonly found genera in the studies, having been identified in 70 and 60% of the publications, respectively. This is interesting, as both have human and plant pathogenic species (e.g., Thomma, 2003; Rivas and Thomas, 2005; Mamgain et al., 2013). Another fungal genus of importance is Fusarium, which encompasses a wide variety of species}, known not only as plant pathogens, but also for their ability to produce mycotoxins and to be opportunistic pathogens for humans (Ma et al., 2013). Additionally, other less represented genera that are also potentially plant pathogens were identified, such as Acremonium, Bipolaris, Colletotrichum, Epicoccum, and Helminthosporium, among others.
In addition to their importance as potential human and plant pathogens, some of these genera stand out for their importance as allergens. Among the main genera identified and reported as potential allergens are Alternaria, Aspergillus, Aureobasidium, Cladosporium, Epicoccum, Fusarium, Helminthosporium, Mucor, Nigrospora, Penicillium, and Rhizopus (Burge, 1985; Horner et al., 1995; Kurup, 2003). This suggests a wide distribution of potentially allergenic fungi in Mexico and that they are taxonomically diverse. Nevertheless, there are very few studies on the relationship between the presence of airborne allergens and hospital admissions for allergies (in particular asthma), so it is important to highlight the work by Rosas et al. (1998), in which pollen and fungal spores were identified as potential allergens.
Finally, molds were also found, of which several species of genera widely distributed in the analyzed literature, such as Alternaria, Aspergillus, Fusarium, Mucor, Phoma, Penicillium and Rhizopus, are known to cause food spoilage (Shapaval et al., 2013). The genus Ulucladium is known to spoil in particular nuts, fruits and cereals (Andersen and Hollensted, 2008). Furthermore, the genera Acremonium, Aspergillus, Penicillium, and Ulocladium have been reported as the most common fungi found in moisture-damaged buildings, which could have implications for human health due to their potential allergenicity, besides their role in the degradation processes of building materials (Andersen et al., 2011).
3.4.5. Other eukaryotes identified
Interestingly, in addition to studies focusing on bacteria and fungi, seven publications were also found on other types of airborne organisms, such as algae, amoebae, protozoa, and yeasts (Table IV). All of these investigations exclusively utilized active samplers and culture-based techniques, and their scope has been limited to four states of the Mexican Republic, most of them carried out in Mexico City (50%). It should be noted that several of these studies found genera that contained both free-living species and some that are hazardous to human health (e.g., Acanthamoeba, Bodo, Candida).
Table IV Main genera of other micro-eukaryote identified in the airborne microbiota studies conducted in Mexico (up to June 2023).
| City(s) | Sampler | Approach | Main micro-eukaryote genera identified | Reference |
| Mexico City | BF | Culture techniques | (Clorophyta), (Cyanophyta) | Rosas et al., 1987 |
| Mexico City | Impringer, EF | Culture techniques | Acanthamoeba, Naegleria | Rivera et al., 1987 |
| Mexico City | Impringer | Culture techniques | Bodo, Cercobodo, Helkesimastix, Monas | Rivera et al., 1992 |
| Mexico City | Impringer | Culture techniques | Acanthamoeba, Hartmannella, Vahlkampfia | Rivera et al., 1994 |
| Minatitlan | BF | Culture techniques | (Clorophyta), (Cyanophyta) | Rosas et al., 1987 |
| Morelia | Vacuum | Culture techniques | Candida, Rhodotorula | Rivera-Tafolla et al., 2022 |
| San Luis Potosí | Impringer | Culture techniques | Acanthamoeba, Hartmannella, Vahlkampfia | Rodríguez-Zaragoza et al., 1993 |
Abbreviations: BF= Bubble flask, EF= Erlenmeyer flasks, Notes: In cases where it was not possible to classify down to the genus level, the lowest taxonomic level is indicated in brackets.
It is worth mentioning that two new species of algae, Brotryokoryne simplex and Chloroglouea microcystoides, were discovered in Mexico City in one of these works (Rosas et al., 1987).
Another interesting finding is that only one of the 35 publications studied airborne pollen. This is probably because pollen is usually considered a bioparticle and is not a microorganism. In the study by Ortega et al. (2019), in addition to investigating airborne fungi, airborne pollen was also examined. They found that the plant taxa that contributed the most pollen to the air were Poaceae, Nyctaginaceae, Ambrosia, and Urticaceae. It is worth mentioning that in addition to this study, there are some other publications on airborne pollen in Mexico (e.g., Tarragó, 1996; Calderón-Ezquerro et al., 2016; Ríos et al., 2016).
The study of other eukaryotes such as algae, protozoa, and amoebae has focused mainly on temperate climate regions, and only few attempts have been made to determine correlations with environmental variables such as solar radiation, relative humidity, temperature, and wind speed. In the case of algae, their abundance has been found to be related to wind speed, vapor pressure, and temperature (Rosas et al., 1989), while the higher abundance of protozoa has been linked to temperature and wind speed and direction (Rivera et al., 1992). Finally, studies on amoebae indicate higher abundance during the dry season and their association with wind speed and direction, and low relative humidity (Rodríguez-Zaragoza et al., 1993).
4. Conclusions and final remarks
The research on airborne microbiota has garnered significant global attention over the past decades, owing to its profound implications for human health and the integrity of agricultural products, structures, and foodstuffs. In Mexico, airborne microbiota investigations have spanned 37 years, culminating in 35 scientific publications. Until 2017, these studies primarily relied on culture or microscopy techniques, with a shift towards metagenomic approaches only starting in 2018. The majority of this work focused on pathogenic microorganisms, and their explorations were predominantly confined to densely populated urban centers.
This comprehensive review identifies several knowledge gaps in Mexico’s understanding of airborne microbiota, highlighting promising areas for future research. The following key aspects merit attention:
Embracing molecular analysis techniques: future studies should adopt molecular analysis techniques, such as metagenomics, to capture a broader spectrum of microorganisms. Alternatively, an interdisciplinary approach that combines multiple methods is advocated, recognizing metagenomics’ limitations in reliably identifying microorganisms beyond the genus level. Adopting molecular techniques will also broaden the spectrum of studied microorganisms, transcending the exclusive focus on pathogenic species.
Analyzing more culturable airborne microorganisms: to talk about pathogenic species it is necessary to characterize phenotypically and molecularly each species, even down to the strain level, as well as to obtain intra- and extra-mural airborne CFU m-3 concentrations, and to calculate the people exposure. This will give a better idea of the potential risk that they pose. The same applies to those microorganisms that have the ability to form freezing nuclei, whose isolation is necessary since this may result in biotechnological applications, e.g., spraying such bacteria to prevent plants from freezing at lower temperatures.
Expanding research sites: the limited exploration of sites hampers the analysis of spatial trends and distribution patterns of airborne microorganisms. Attention must extend beyond urban centers to encompass diverse regions, including deserts, mountains, forests, small rural human settlements, and other intriguing sites.
Significance of agricultural area analysis: routine analysis of airborne microbiota in agricultural regions assumes critical importance as it can unveil crop pathogens, facilitating early disease outbreak prevention and thus safeguarding agricultural productivity.
Correlation analyses for enhanced insights: thorough correlation analyses incorporating factors such as altitude, geography, climate, biological characteristics of the region, and pollutant levels, will offer nuanced insights into the structure and distribution of airborne microorganism communities.
Unveiling temporal dynamics through longitudinal analyses: longitudinal studies are essential to elucidate the behavior of airborne microbial communities over time. These investigations should consider changes in air composition, as well as in physical parameters such as temperature and UV irradiance, which are necessary to understand the temporal shifts in microbial communities.
Exploring abiotic particle interactions: to advance our understanding of the ecology and behavior of the airborne microbiome in Mexico, comprehensive investigations into correlations between airborne microorganism communities and the physical and chemical characteristics of airborne abiotic particles are indispensable.
This comprehensive review serves as a foundation for further research endeavors, aiming to enhance Mexico’ss understanding of airborne microbiota. By integrating state-of-the-art techniques, exploring diverse environments, and fostering interdisciplinary collaboration, the scientific community can unlock groundbreaking discoveries in this crucial field.











 nueva página del texto (beta)
nueva página del texto (beta)





