Introduction
Diatoms are an important and diverse group of organisms that fix almost 20% of global carbon (Armbrust, 2009; Malviya et al., 2016; D. G. Mann, 1999; Onuma et al., 2017). However, aside from the toxic diatoms, the taxonomy of the specious class of Bacillariophyceae is ‘in a rather poor state’ (Mann et al., 2020). The taxonomic history of the genus Halamphora has been dynamic since its revival by Levkov (2009) produced a considerable number of inclusions via transfers of species new nomenclatural combinations, mostly from the genus Amphora, descriptions of new species, and taxonomic validations. The increased number of taxonomically accepted species and intraspecific names registered in Algaebase is remarkable, from 110 in 2018 to 163 in 2023 (Guiry & Guiry 2023), which reflects the degree of complexity that this genus still represents.
In diatom, monoclonal cultures form a vital part of an integrative taxonomic approach to identifying species using morphological and molecular data (Mohamad et al., 2022). Also, electron microscopy of monoclonal cultures is commonly used to describe new diatom species with ultrastructural details. Stepanek & Kociolek (2018) described H. adumbratoides and complemented their description with molecular tools. Electron microscopy and molecular information have contributed significantly to the determination of new species of diatoms, taxonomic validations, and the transfer of taxa from one genus to another. For example, taxonomic relocations of taxa have been carried out with the closely related genera Amphora, Cymbamphora, and Seminavis (Levkov, 2009; Stepanek & Kociolek, 2013; 2015, 2018, 2019; Van de Vijver et al., 2014).
Morphometric similarity between taxa of different genera is rare; however, it is not uncommon among species of closely related genera. For example, the morphology of H. adumbratoides is very similar to that of Amphora adumbrata in terms of the shape of the valve and the fine striae composed of a single elongated areola (Stepanek & Kociolek, 2018). The number of striae was a key characteristic for differentiating these taxa by traditional taxonomy (morphology) (Desianti et al., 2015; Stepanek & Kociolek, 2018). At the molecular level, SSU, rbcL, and psbC markers were used to identify H. adumbratoides (Stepanek & Kociolek, 2018).
In geographical terms, H. adumbratoides has been recorded only from the coasts of the Atlantic Ocean, in Florida Bay, Cotton Key, Monroe Country, FL, USA. In this publication, we present morphological characteristics and use molecular identity of this taxon (strain CIBA 160) to report its presence in the Pacific Ocean region from the coastal Balandra lagoon (La Paz Bay) in the Gulf of California, on the southern coast of the Baja California Peninsula, Mexico. It is the second recording but the first interoceanic registration of H. adumbratoides. We also register considerable morphological features and morphometry variations in the laboratory-cultured cells.
Materials and methods
Sample collection and morphological observation
Water samples of 50 mL were taken by triplicate from the Balandra lagoon (24°19′9.01′′N 110°19′18.17′′ W) near La Paz, Baja California Sur (Figure 1), in December 2018, and brought to the laboratory. The samples were filtered through a 30-μm Nitex mesh and f/2 medium with silicates added to the filtered samples (Guillard, 1975).
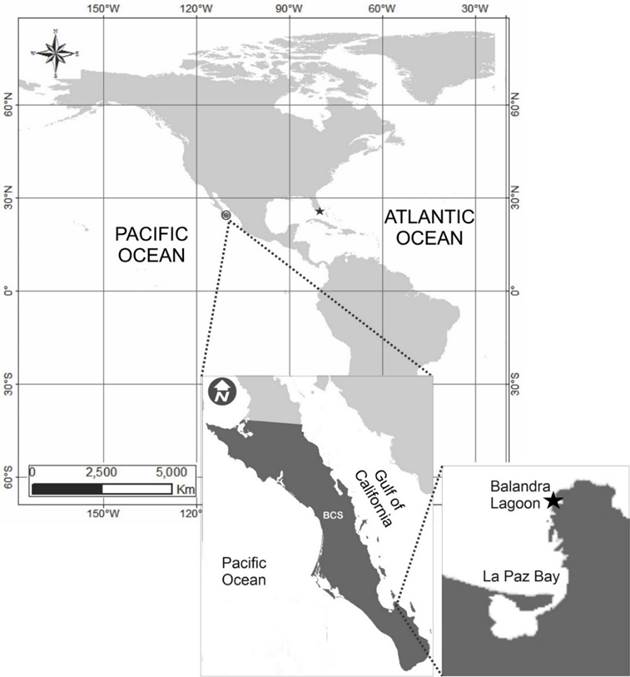
Figure 1 Current recordings of Halamphora adumbratoides in the Pacific and Atlantic Ocean (solid stars). Inset figures show an expanded map of the collection site in the Balandra Lagoon, Mexico.
After two weeks, microalgal cells were isolated in a 1:50 dilution in f/2 medium and streaked on marine agar plates containing the same medium. Single-cell colonies were isolated under low magnification in a microscope, with a sterile hypodermic needle and then transferred to 24-well plates containing 2 ml of culture medium. Two weeks later, the cells were transferred to test tubes with 5 ml of culture medium, and the unialgal culture of isolated cells was examined under light microscopy. Once confirmed, the monoclonal cultures were deposited in CIBNOR’s Microalgae Collection under catalog number CIBA 160 (https://www.cibnor.gob.mx/investigacion/colecciones-biologicas/coleccion-de-microalgas) after three reseeding periods. We have a collection of three fixed slides with the skeletons of the cells, that remains in the laboratory.
For light and electron microscopy and molecular analyses, cells were cultured in triplicate in 500-mL flasks with 250 mL of medium at 25±1°C, 40 μmol·m-2·s-1, 12:12 h, and 35 PSU. At the exponential stage (on the sixth day), separate samples were taken for each analysis. Live and cleaned cells were observed under a Zeiss Axio Lab A1 microscope, and photographs were taken with a Canon EOS Rebel T5i camera (Figure 2). The cells were prepared for scanning electron microscopy (SEM) as indicated previously by López-Fuerte et al. (2020). Briefly, the organic matter was eliminated by oxidation, heat-assisted with concentrated nitric acid. The cells were then washed in distilled water to reach a neutral pH. SEM images of the cells were taken with a Hitachi SU3500 electron microscope operating at 10 kV and a 6-mm working distance. A coverslip holding the cells was attached to a 32-mm aluminum stub using conductive carbon tape and coated with around 15 nm of gold in a Hummer 6.2 sputtering unit.
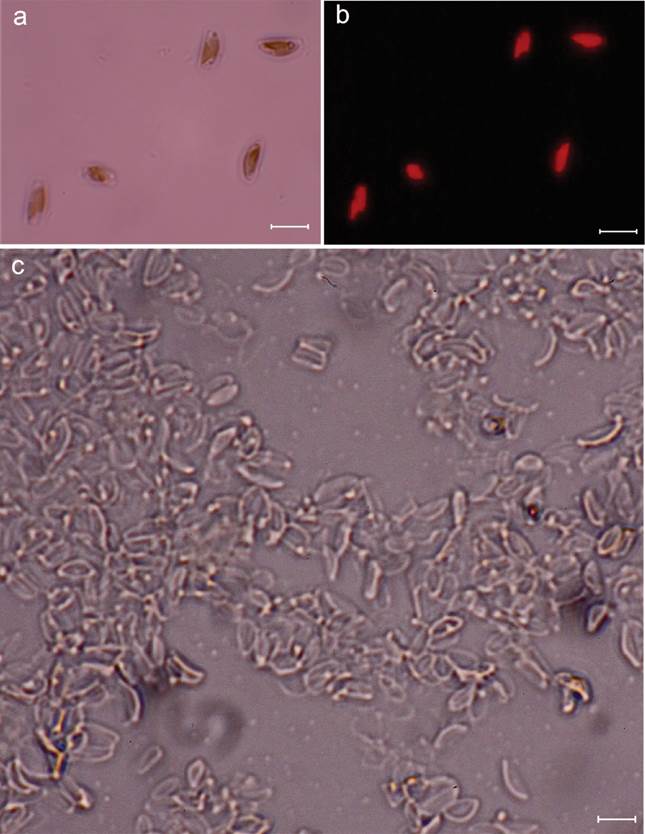
Figure 2 a-c. Halamphora adumbratoides, a. live cell showing plastids; b. fluorescence microscopy showing plastids; c. cleaned specimens. Scale bars represent 5 μm.
The key morphological characteristics -length, width, and density of the striae- were measured by light and electron microscopy and compared to those reported for Halamphora adumbratoides (Stepanek & Kociolek, 2018). The validity nomenclatural status of the name was verified on the Algaebase website (Guiry & Guiry 2023).
DNA extraction, PCR amplification, and sequencing
Cells of the CIBA 160 strain obtained from the exponential stage cultures were utilized to extract genomic DNA by the CTAB method (Doyle, 1991; Herrera et al., 2014). Four regions of DNA were analyzed: fragment of two nuclear genes, the small sub-unit of nuclear gene 18S ribosomal RNA (SSU), and the large subunit of nuclear gene 28S ribosomal RNA (LSU) were amplified following the PCR conditions described in López-Fuerte et al. (2020). A partial sequence of the ribulose bisphosphate carboxylase/oxygenase gene (rbcL) from the chloroplast was amplified according to the published protocols (López-Fuerte et al., 2020). Also, the photosystem II CP43 protein gene (psbC) was amplified with 30 picomoles of the psbC+ and psbC- primers (Alverson et al., 2007) in 40 μL of a PCR reaction mixture containing genomic DNA, 0.3 mM dNTPs, 2 mM MgCl2, 4 μL of 10X PCR buffer, and 2 units of platinum Taq polymerase (InvitrogenTM).
The PCR conditions for psbC amplification were denaturation at 94°C for 3.30 min, then 35 cycles of 30 s at 94 °C, 50 s at 52 °C, and 80 s at 72 °C. The final extension was for 10 min at 72 °C. Both strands of all markers were sequenced with the amplification primers and an internal primer for the SSU and chloroplast genes (18S 962R, rbcL1255, psbC857).
We edited the four sequences (SSU, LSU, psbC, and rbcL) in DNA Baser 4.5 program (http://www.dnabaser.com) and compared the SSU, rbcL, and psbC sequences to those of H. adumbratoides in the GenBank database. The phylogenetic relationship of the CIBA 160 strain was inferred based on the concatenated nucleotide sequences of SSU, LSU, psbC, and rbcL with 23 Halamphora species (Table 1) using the maximum likelihood (ML) method in PAUP 4.0a166 (Swofford, 2002) and Bayesian inference (BI) in MrBayes 3.2.7a (Ronquist et al., 2012). The best fit nucleotide model TIM3+I+G selected according to the BIC criterion in jModeltest 2.1.10 (Darriba et al., 2012; Guindon & Gascuel, 2003) for the concatenated nucleotide sequences was implemented in both analyses. Maximum likelihood analysis was performed with 500 pseudoreplicates under the option of heuristic tree-searching with tree bisection-reconnection branch swapping to generate a majority rule ML consensus tree. Bayesian inference was run for 10 million generations, and a majority rule BI consensus tree was obtained after eliminating 25% of the initial trees using the burn-in option.
Table 1 Halamphora and Amphora sequences used in the phylogenetic analyses.Tabla 1. Secuencias de Halamphora y Amphora utilizadas en los análisis filogenéticos.
| Taxa | 18S | rbcL | psbC | 28S | Reference |
|---|---|---|---|---|---|
|
Halamphora adumbratoides AMPH041 |
MG027270 | MG027434 | MG027514 | NA | Stepanek & Kociolek, 2019 |
|
Halamphora adumbratoides CIBA 160 |
ON714546 | ON736839 | ON736840 | ON714544 | This study |
|
Halamphora pellicula AMPH134 |
MG027316 | MG027481 | MG027561 | MG027401 | Stepanek & Kociolek, 2019 |
|
Halamphora pellicula AMPH153 |
MG027320 | MG027486 | MG027566 | MG027407 | Stepanek & Kociolek, 2019 |
|
Halamphora elongata AMPH001 |
MG027259 | MG027423 | MG027503 | MG027337 | Stepanek & Kociolek, 2019 |
|
Halamphora aponina AMPH049 |
MG027275 | MG027439 | MG027519 | MG027355 | Stepanek & Kociolek, 2019 |
|
Halamphora aponina AMPH102 |
MG027296 | MG027461 | MG027541 | MG027381 | Stepanek & Kociolek, 2019 |
|
Halamphora pseudoholsatica AMPH165 |
MG027327 | MG027493 | MG027573 | MG027414 | Stepanek & Kociolek, 2019 |
|
Halamphora holsatica AMPH154 |
MG027321 | MG027487 | MG027567 | MG027408 | Stepanek & Kociolek, 2019 |
|
Halamphora rushforthii AMPH117 |
MG027306 | MG027471 | MG027551 | MG027391 | Stepanek & Kociolek, 2019 |
|
Halamphora coffeaeformis AMPH104 |
MG027297 | MG027462 | MG027542 | MG027382 | Stepanek & Kociolek, 2019 |
|
Halamphora pratensis AMPH106 |
MG027299 | MG027464 | MG027544 | MG027384 | Stepanek & Kociolek, 2019 |
|
Halamphora isumiensis AMPH164 |
MG027326 | MG027492 | MG027572 | MG027413 | Stepanek & Kociolek, 2019 |
|
Halamphora tenucostata AMPH042 |
MG027271 | MG027435 | MG027515 | MG027350 | Stepanek & Kociolek, 2019 |
|
Halamphora tenuis AMPH034 |
MG027269 | MG027433 | MG027513 | MG027348 | Stepanek & Kociolek, 2019 |
|
Halamphora scatebra AMPH119 |
MG027308 | MG027473 | MG027553 | MG027393 | Stepanek & Kociolek, 2019 |
|
Halamphora subturgida AMPH015 |
MG027260 | MG027424 | MG027504 | MG027338 | Stepanek & Kociolek, 2019 |
|
Halamphora bicapitata AMPH055 |
MG027278 | MG027442 | MG027522 | MG027359 | Stepanek & Kociolek, 2019 |
|
Halamphora nagumoi AMPH166 |
MG027328 | MG027494 | MG027574 | MG027415 | Stepanek & Kociolek, 2019 |
|
Halamphora oligotraphenta AMPH009 |
KJ463451 | KJ463481 | KJ463511 | KP229528 | Stepanek & Kociolek, 2014; Stephanek et al. 2015 (28S) |
|
Halamphora veneta AMPH005 |
KJ463452 | KJ463482 | KJ463512 | KP229530 | Stepanek & Kociolek, 2014; Stephanek et al. 2015 (28S) |
|
Halamphora venetoides AMPH017 |
KJ463453 | KJ463483 | KJ463513 | NA | Stepanek & Kociolek, 2014, 2018 |
|
Halamphora coloradiana AMPH025 |
KJ463450 | KJ463480 | KJ463510 | KP229529 | Stepanek & Kociolek, 2014; Stephanek et al. 2015 (28S) |
|
Amphora allanta AMPH129 |
MG027314 | MG027479 | MG027559 | MG027399 | Stepanek & Kociolek, 2019 |
|
Amphora commutata AMPH126 |
KP229526 | KP229547 | KP229549 | KP229545 | Stepanek et al., 2015 |
Results
The length and width recorded for the valves of the laboratory cultured cells of the strain CIBA 160 (Halamphora adumbratoides) are 4.1-6.6 and 1.6-2.4 μm, respectively. The number of dorsal and ventral striae for the CIBA 160 strain are 57-60 and 60-70 in 10 μm, respectively. The valve shape is semi-elliptical, and the ends are broadly rounded (Figure 3a -j). The raphe is straight with straight proximal ends, while the distal raphe ends are deflected to hooked dorsally (Figure 3a-b). Externally, two pores are visible at the end of the dorsal raphe ledge at each end of the raphe (Figure 3a-d black arrow), or they may appear fused (Figure 3d white arrow), while internally two, only a single pore is observed (Figure 3e-g, j). Internally, the proximal raphe ends terminated in a broad, fused central helictoglossa (Figure 3e-f, h, white asterisk). The dorsal raphe ledge is nearly absent, especially through the valve center (Figure 3a-d). A reduced dorsal raphe ledge is present, more accentuated towards the distal end of the valve (Figure 3a-b, black asterisk). The dorsal and ventral striae are continuous through the valve center, slightly radiate (Figure 3a-d), and not resolved in the LM (Figure 2b). Figure 3e-j shows internal views of the deformed valves and other deformed structures, like the raphe, which is in a different position than in the undeformed valves.
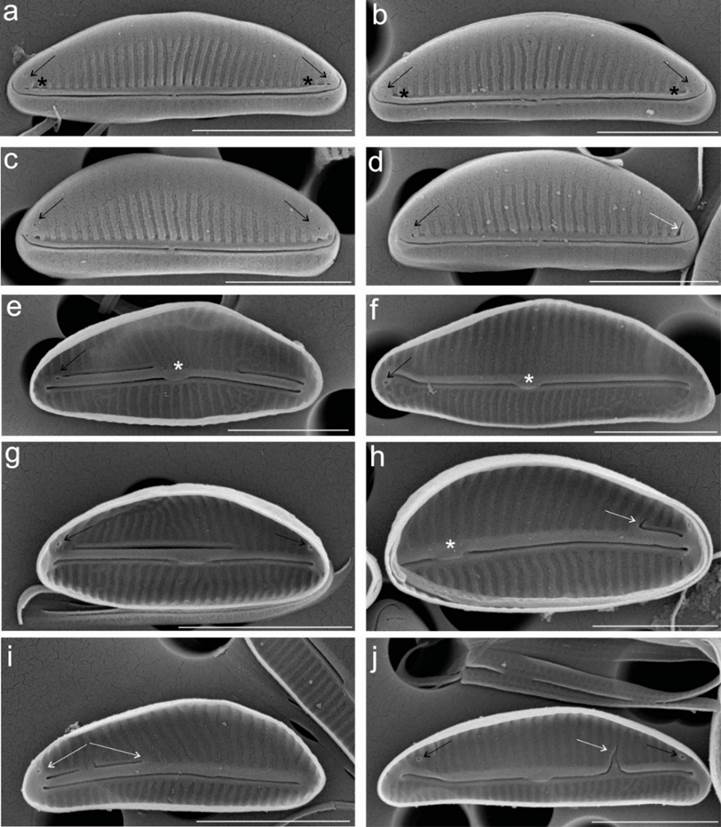
Figure 3 a-d. External view of the valve. a-d showing two distal pores (black arrows). The external and internal proximal raphe ends are not deflected, but the external distal raphe ends are dorsally deflected, a-d, e. Externally, the dorsal and ventral raphe ledges are small but continuous along the length of the valve, e-j. Internal view of the valve. e, f, h. Internally, the proximal raphe ends terminate in a broad, fused central helictoglossa (white asterisks), e. f, g, h, j. One distal pore is visible (black arrows). f, g, h, i, j. White arrows show the abnormal dorsal raphe ledge. Scale bars correspondence: a, 3 μm, b-j, 2 μm.
Molecular analysis
We obtained 4,004 bp nucleotide sequences (SSU 1375 bp, rbcL 1444 bp, psbC 1185 bp; GenBank accession numbers: ON714546, ON736839, ON736840) for the CIBA 160 strain (H. adumbratoides) from the Pacific Ocean. Comparison of our sequences to those of the type species from the Atlantic Ocean in GenBank showed 100, 99.8, and 99.5% similarity for SSU, rbcL, and psbC, respectively. The BI and ML phylogenetic analyses placed the H. adumbratoides strains in one clade with H. pellicula as a sister clade (Figure 4). This node was supported strongly by the BI (100%) and ML (92%) analyses.
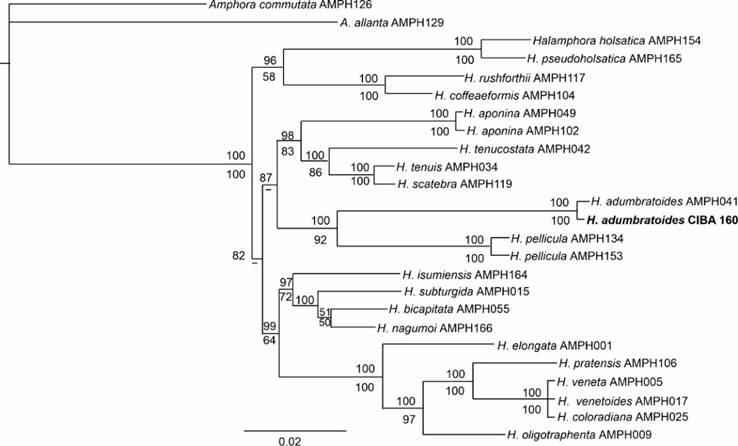
Figure 4 Bayesian tree of the Halamphora species based on the partial sequences of SSU, LSU, psbC, and rbcL. Bootstrap values of maximum likelihood analysis are given below, and posterior probabilities of Bayesian inference are given above the nodes. Nodes supported by less than 50% of Bootstrap values are indicated with the symbol ˋ-ˊ
Discussion
Accurate identification of a species depends, in principle, on the methods' reliability. Historically, and even today, the identification of a diatom species is based on morphometric aspects observed under light microscopy, which requires extensive taxonomic experience (Mora et al., 2019). The characterization of certain diatom species is impossible only with this tool because of their small size and inconspicuous diagnostic structures (Li et al., 2018; D. Mann et al., 2020). Fortunately, this issue can be resolved with electron microscopy. However, not all diatom species can be delineated using morphology, especially species with subtle morphological differences (Beszteri et al., 2007; Evans et al., 2007; Malviya et al., 2016). To aid in determining species boundaries among diatoms, molecular tools, such as DNA sequences are applied (An et al., 2017; Beszteri et al., 2007; Evans et al., 2007; Guillard, 2005; D. Mann et al., 2010, 2020; Medlin, 1991). However, this methodology has limitations as it may be unable to resolve the species identity (Malviya et al., 2016) without conspecific and/or closely related species sequences in the nucleotide database. Therefore, morphological, and molecular methods are important in diatoms and should be considered as elements of a synergistic approach to species identification challenges.
After a morphometric review of the images of the laboratory cultured cells of the CIBA 160 strain obtained with both light and electron microscopy, it was not possible to identify this strain with any previously described taxon. However, approximations were made with Amphora adumbrata and Halamphora adumbratoides. Nevertheless, the morphometric analyses did not allow the taxonomic determination of the CIBA 160 strain as H. adumbratoides because the morphology of this strain does not exactly match the original description reported by Stepanek & Kociolek (2018), specifically due to the shape and size of the valve. In the original description of H. adumbratoides, the valves were narrow and semi-lanceolate, and the valve ends narrowly rounded to weakly subcapitate in larger specimens. The dorsal and ventral margins, however, do coincide with the original description, as they are arched and straight, respectively (Figure 3a-d). The presence of pores at the end of the dorsal raphe ledge at each end of the raphe was not mentioned in the original description. The axial area, not easily distinguishable in the original description, was not observed in the specimens of the CIBA 160 strain analyzed.
For the length and width of the valves, Stepanek & Kociolek (2018) reported ranges of 8.0-10.0 and 1.5-2.5 μm, while the cells (N=24) of the CIBA 160 strain recorded valve lengths and widths of 4.1-6.6 and 1.6-2.4 μm. Only the length range differed, as it was much shorter than in the original description. The number of dorsal and ventral striae in the CIBA 160 strain almost coincided with the original description, with 56-57 dorsal striae and ca. 70 ventral striae in 10 μm in the original description, while the striae range in 10 μm for the CIBA 160 strain were 57-60 and 60-70 for the dorsal and ventral striae, respectively.
A possible explanation for the size differences and morphological changes could be the effect of laboratory culturing on the cells of CIBA 160. In diatoms Rose & Cox (2013), Petrova et al. (2020) and Mohamad et al. (2022) have reported morphological changes in laboratory cultured cells. Diatoms tend to produce small-sized cells over time due to asexual reproduction (Mohamad et al., 2022). This phenomenon has been demonstrated in 15 monoclonal pennate diatoms, but the width and number of striae did not differ from the original description (Mohamad et al., 2022). Our results support the findings of Mohamad et al. (2022), and their proposal that the number of striae could be a useful character for species discrimination.
Our molecular analyses confirmed that the CIBA 160 strain belongs to H. adumbratoides by showing very high similarity to the sequences of this species from the Atlantic Ocean: 100, 99.8, and 99.5% similarity for SSU, rbcL, and psbC, respectively. The present study corroborates the results of Mohamad et al. (2022) that molecular data remain consistent irrespective of morphological changes in laboratory-cultured diatoms. Phylogenetic analyses of this study were also congruent with the relationship presented by Stepanek & Kociolek (2019) for the H. adumbratoides species, which is a sister group to H. pellicula.
We have registered morphological abnormalities in the cells of CIBA 160. The helictoglossa (Figure 3h white asterisk) is displaced to the left side of the valve. In contrast, the dorsal raphe ledge is very short and located only to the right of the valve (white arrow) (Figure 3h). The exact opposite configuration is visible in Figure 3i (white arrow), where the structure is seen to the left of the valve. These abnormalities could be associated with long-term culture of cells in the laboratory (Estes & Dute, 1994; Falasco et al., 2009; Petrova et al., 2020).
In the genera Amphora and Halamphora, most taxonomic determinations and descriptions of new species are based on morphological analyses of specimens from natural populations (Levkov, 2009; Wachnicka & Gaiser, 2007). However, laboratory-cultured cells were also used for integrative taxonomic studies, combining morphological and molecular data (López-Fuerte et al., 2020; Stepanek et al., 2013, 2015a, 2015b, 2018).
It is important to mention that regardless of the origin of diatoms (natural or cultivated), the morphological description of a species must be accurate and informative, based on as much information as possible. This is because diatoms may present morphological variations throughout their life history, whether in the natural or artificial environment (Rose & Cox, 2013).
In our study, using morphological and molecular data has resulted in the reliable taxonomic identification of the CIBA 160 strain. Our results support using an integrative approach for fine-grained integrative taxonomic studies and the importance of laboratory cultures in the diatom taxonomy (Mohamad et al., 2022).
Conclusions
The taxonomic identification of the CIBA 160 strain as H. adumbratoides was not possible based only on morphometric characteristics. This was due mainly to differences in the valve leaflet length and the presence of pores at the extremes of the valve on the raphe channel. Molecular analyses, however, allowed us to determine species identity with reference DNA sequences deposited in GenBank. Using DNA sequence data is important for cryptic, pseudo-cryptic diatom species and diatoms in general. Integrated approaches can play a vital role in discovering species with broad morphological variations.











 nueva página del texto (beta)
nueva página del texto (beta)



