Introduction
Lignocellulosic material, and specifically wood, is naturally degraded by the action of fungi, mainly white-rot basidiomycetes such as Trametes, Phanerochaete, Ganoderma, Coriolus, Phlebia, Pycnoporus, Mycoacia, Hyphodontia, Bjerkandera, Stereum, Hypodermella, Gymnopus, Laetiporus, Laetiporus Stereum, Fomitiporella, Dacrymyces, Sistotrema, Penioporella, and Entoloma among others (Ríos and Eyzaguirre, 1992; Ortiz et al., 2014). Its mechanism of action implies the secretion of hydrolytic enzymes such as cellulase, xylanases, and oxidative enzymes such as laccase, lignin peroxidase, and manganese peroxidase (Arana et al., 2002).
The basidiomycete fungi do not act alone; Blanchette and Shaw (1987) proposed the association of bacteria (Enterobacter spp.) and yeasts (Saccharomyces bailii and Pichia pinus) with basidiomycete fungi that contribute to the complete mineralization of the wood. Several studies have been conducted to understand microbial interactions during wood degradation. One of the processes studied is the wood degradation known as “Palo podrido”, being the main microorganism responsible for this process the basidiomycete fungi, Ganoderma applanatum. This degradation is located exclusively in the south of the American continent, specifically in the Ulmo tree (Euryphia cordifolia).
Various microorganisms associated with wood degradation processes have been isolated using plate or liquid dilution techniques. Some genera of yeasts with xylanolytic activity, such as Kloeckera, Pseudozyma, Kodamaea, Pichia, Asterotremella, Sporidiobolus, Ogataea, Spencermartinsiella, Saturnispora, Galactomyces, Bulleromyces, Lindomy, Saturnispocesella, Scheffomersyia, Saturnispocesellay, Cryptococcus, Trichosporon, and Candida have been isolated (Morais et al., 2013; Lara et al., 2014). Similarly, yeasts with cellulolytic activities have been reported, including Kloeckera, Rhodotorula, Debaryomyces, Guehomyces, Hanseniaspora, Dioszegia, Cryptococcus, Pichia, Trichosporon, Arthroascus, and Candida (Brandão et al. 2011; Sarawan et al., 2013; Bautista-Guerrero et al., 2022). However, the reports of laccase-activity have only been reported for Cryptococcus spp. yeast (Chen and Williamson 2011) and Yarrowia (Lee et al., 2012).
The use of yeasts with the capacity to produce laccase has advantages compared to filamentous fungi, such as easy handling of cells, more economic substrates for their cultivation, rapid growth, and easy genetic manipulation, in addition to carrying out post-translational modifications such as glycosylation, which facilitates the expression of active enzymes (Karbalaei et al., 2020; Putra et al., 2022). On the other hand, the lignocellulolytic activity may depend on the growth medium of the yeast. There are few reports on studies of the effect of the culture medium on the production of lignocellulosic enzymes in yeast (Lara et al., 2014). This factor is important if the industrial potential of yeast is considered, as is the case of cellulases and xylanases of great economic impact because these enzymes are commonly used in textile industry, food industry, pulp, paper industry, agricultural area, and ethanol production, among others (Johnson 2013; Phitsuwan et al., 2013).
This study aim was to isolate and identify yeasts present in the wood degradation of Euryphia cordifolia known as “Palo podrido”, by evaluating different carbon and nitrogen sources and selecting the strains with cellulolytic, xylanolytic, and laccase activities. In addition, it will allow a better understanding of this degradation type of great environmental importance due to its participation in the carbon cycle of the planet.
MATERIALS AND METHODS
Yeast isolation
Yeasts were isolated from Eucryphia cordifolia wood with symptoms of degradation known as “Palo podrido”. The wood samples were collected from the Valdivian rainforest in the tenth region of Chile (Isla Grande de Chiloé: 42°01’20.05’’ S, 74°01’19.03’’ W), in the winter season. One Gram of the collected samples was homogenized in 10 mL of peptone water, and serial dilutions were made. YEPD agar plates (DifcoTM) supplemented with ampicillin (100 mg/L) were inoculated with the dilutions obtained and incubated at 28 °C for 72 h; the colonies were purified by successive reseeding on YEPD agar and kept at 4 °C on a plate until their use. Yeasts from a 72 h culture in YEPD were stored at - 20 °C in 2 mL cryotubes containing 25 % glycerol and 75 % YEPD broth (Avchar et al., 2022).
Molecular identification
The isolated yeasts were cultured in YEPD broth at 28 °C in an orbital shaker at 150 rpm for 72 h. Cell cultures were centrifuged at 13,000 x g for 3 min. After separating the supernatant, the pellets were washed with 1.5 mL of sterile water and centrifuged again. The final pellet was added with the following specifications: 0.3 g of sterile glass beads with 500 µL of a solution of 10 mM HEPES, 0.5 mM sucrose, 20 mM EDTA, and pH 6.9. Each sample was vigorously vortexed for 1 min, followed by 1 min on ice; the process was repeated six times. Subsequently, the DNA extraction protocol of Raeder and Broda (1985) was followed. The concentration and integrity of the DNA were confirmed with a nanodrop spectrophotometer (Nanodrop 2000, Thermo Scientific) and 1 % electrophoresis gel in a 1X TAE buffer (100 V, 1 h), respectively.
The ITS region of each yeast was used as a molecular marker. The ITS region was amplified using the universal primers ITS4 (5’TCCTCCGCTTATTGATATGC3 ‘) and ITS5 (5’GGAAGTAAAAGTCGTAACAA3’) (White et al., 1990). Fifty microliters of the reaction mix contained 1X PCR buffer, 200 µM of each dNTP, 2 mM of MgCl2, 10 pmol of each primer, 1-5 ng of DNA, and 1 U of Taq DNA polymerase. The amplification reaction was carried out in a thermal cycler (SimpliAmp, Thermal Cycler, Applied Biosystems) under the following conditions: 5 min of initial denaturation at 95 °C, followed by 30 denaturation cycles at 95 °C for 45 sec, alignment at 52 °C for 45 sec, and extension at 72 °C for 45 sec, with 5 min of final extension at 72 °C. The amplified products were purified with the GenElute PCR Clean-up Kit (Sigma). The fragments were sent to the Institute of Cell Physiology, UNAM (Mexico) Molecular Biology Unit for sequencing. The originated electropherograms were analyzed using the BioEdit Sequence Alignment Editor software (version 7.1.11) and Chromas Lite (version 2.1). The sequences were subjected to a similarity analysis with the BLAST tool of the NCBI portal.
Phylogenetic analysis
The selected sequences for the phylogenetic analysis were aligned with the ClustalX Program version 2.1, using the parameters gap opening: 75 and gap extension and 3.3 for multiple alignments. Later, they were visually revised and optimized. The regions with ambiguous alignments were removed, and the length was uniform by eliminating the extremes. The BEAST v.1.5.4 platform was used to perform the phylogenetic analysis through Bayesian inference using a Monte Carlo Markov Chains (MCMC) algorithm.
The nucleotide substitution model selected and optimized by the Akaike information criterion (AIC) for phylogenetic inference was GTR, obtained using the jModelTest 2 software. The molecular clock that best fit the data was selected by Bayes comparison Factor (BF). After removing 10 % of the burning, the tree with the maximum credibility (MCC) was obtained by agreeing to 10,000,000 trees using the Tree Annotator software v.1.5.13. The phylogenetic tree was visualized and rooted in FigTree v1.2.2. All software used for molecular analysis is freely available.
Assays of carbohydrate assimilation
Three mL of YNB broth (Thermo Scientific) inoculated with each strain (0.3 mL, A600 = 1) and supplemented with 0.2 % of each carbohydrate tested in 15 mL tubes were used. The carbohydrates tested were glucose, fructose, galactose, cellobiose, and xylose (Qadri and Nichols 1978). Each tube was incubated at 28 °C and 150 rpm for 2 days. The tests were carried out in triplicate. The positive assimilation of the carbohydrate was qualitatively evidenced by the presence of turbidity in the YNB broth (Thermo Scientific), reported as growth (+) or no growth (-). On the other hand, for the qualitative detection of gas production (CO2), the protocol described by Brooks (2008) was used.
Assays of cellulolytic and xylanolytic activity in plate
Ten microliters of each yeast strain were inoculated in 30 mL of YEPD broth (Thermo Scientific) and incubated at 28 °C in an orbital shaker at 150 rpm for 72 h. The pre-inoculum was prepared by adjusting the cell culture to A600 = 1 in peptone water. YNB agar plates supplemented with 0.2 % birch xylan were prepared to determine xylanolytic activity. YNB agar plates supplemented with 0.2 % CMC (carboxymethylcellulose) were prepared for cellulolytic activity.
The growth of the microorganism was determined by measuring the diameter of the colony. The plates were revealed by adding 10 mL of Gram’s iodine solution (1: 5) to the culture surface and incubating for 5 min at room temperature. The production of xylanases and cellulases was recorded by observing a clear area around the colony (Kasama et al., 2008); only the difference between the hydrolysis halo and the colony size in triplicate of the yeasts under study was determined. The halo was measured to calculate the power index (PI) according to the formula (Cruz-Ramírez et al., 2012):
Effect of nitrogen source on microbial growth
The strains with the highest hydrolytic activity were inoculated in grown media with different Nitrogen concentrations to evaluate their effect on cellulase, xylanase, and laccase production. Birch xylan (0.2 %), and CMC (0.2 %) were used to detect xylanolytic and cellulolytic activities, respectively. The Nitrogen sources tested were Casein Peptone-H (Bioxon) (CP-H), Casein Peptone-H (MCD LAB) (CP-H2), Proteose Peptone No. 3 (BD DIFCO) (PP-3), Bacto Tryptone (DIFCO) (BT), and Gelatin Peptone (DIBICO) (GP). The concentrations tested were 1, 2, and 3 % of each Nitrogen source studied.
All tests were performed in Petri dishes with 1.5 % bacteriological agar by triplicate. The total concentration of each Nitrogen source was determined with a LECO®FP-528 C N2/Protein determiner at the Food Technology Laboratory 2 of the UAEH (Universidad Autónoma del Estado de Hidalgo, México). The PI of each assay was determined as described in the previous section.
Determination of laccase activity
For this study, YNB agar plates supplemented with 0.2 % glucose and ABTS (2,2’-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid)) 2 mM were used. Each plate was inoculated with 10 µL of each yeast strain under study and incubated at 28 °C for 96 h. The enzymatic activity was quantified by measuring PI (Cruz-Ramírez et al., 2012). Subsequently, the effect of the carbon source on the production of this enzyme was evaluated. Plates were prepared with YNB agar supplemented with 2 mM ABTS and glucose, xylose, or glycerol at three different concentrations (0.5, 1, and 2 %). After inoculation, strains were incubated at 28 °C for 2 days.
Results
Isolation and molecular identification of yeasts
A total of ten yeast strains were isolated from the “Palo podrido” sample collected on the island of Chiloé (Chile). The phylogenetic study allowed the identification of 3 strains of Phylum Basidiomycota (Rhodotorula, Cryptococcus, and Leucosporidium) (Figure 1), and seven strains belonging to Phylum Ascomycota (Candida, Sugiyamaella, and Spencermantinsiella) (Figure 2). Table 1 shows the results obtained from molecular identification using the ITS molecular markers and the accession number of the sequences deposited in GenBank. Each isolated strain presented different morphological characteristics, so they were classified by the key to the culture in which they were isolated. The yeasts were tested to determine their monosaccharide assimilation capacity, hydrolytic enzyme production (cellulase and xylanase), and laccase production.
Table 1. Yeast identified from wood degradation known as “Palo podrido” and comparisons with the best BLAST match using the NCBI GenBank database.
Tabla 1. Levaduras identificadas del proceso de degradación de madera “Palo podrido” y comparaciones con el mejor alineamiento en BLAST utilizando la base de datos del GenBank del NCBI.
| Culture | Best BLAST match | E value | Max identity | Gen Bank accession # |
| Basidiomycetous | ||||
| AIRB-ML2.1 | Rhodotorula sp. | 0.0 | 95 | KP326401.1 |
| AIRB-ML4.2 | Cryptococcus sp. | 0.0 | 93 | KP326402.1 |
| AIRB-ML5.1 | Leucosporidium sp. | 0.0 | 99 | KP326403.1 |
| Ascomycetous | ||||
| AIRB-ML1.1 | Candida sp. | 4e-160 | 96 | KP326404.1 |
| AIRB-ML4.1 | Sugiyamaella sp. | 0.0 | 94 | KP326411.1 |
| AIRB-ML5.3 | Sugiyamaella sp. | 0.0 | 99 | KP326410.1 |
| AIRB-ML6.4 | Sugiyamaella sp. | 0.0 | 99 | KP326406.1 |
| AIRB-ML7.1 | Candida sp. | 0.0 | 99 | KP326408.1 |
| AIRB-ML9.2 | Spencermartinsiella sp | 2e-170 | 89 | KP326409.1 |
| AIRB-ML11.1 | Sugiyamaella sp. | 0.0 | 99 | KP326407.1 |
Assimilation
Carbohydrate assimilation tests were carried out to determine the ability of yeasts to assimilate and ferment sugars. The results are shown in Table 2. The results coincided with the data reported by the Fungal Biodiversity Center (CBS-KNAW) (http://www.cbs.knaw.nl/), finding fructose assimilation in all the microorganisms studied as a novelty.
Table 2. Carbohydrate assimilation capacity of yeasts isolated from the degradation of “Palo podrido”.
Tabla 2. Capacidad de asimilación de carbohidratos de las levaduras aisladas de la degradación de “Palo podrido”.
| Carbohydrate | Glucose | Galactose | Fructose | Cellobiose | Mannose | Xylose |
| Rhodotorula sp. AIRB-ML2.1 | + | + | + | + | + | + |
| Cryptococcus sp. AIRB-ML4.2 | + | - | + | + | + | + |
| Leucosporidium sp. AIRB-ML5.1 | + | + | + | + | - | + |
| Candida sp. AIRB-ML1.1 | + | + | + | + | - | + |
| Sugiyamaella sp. AIRB-ML4.1 | + | + | + | + | - | + |
| Sugiyamaella sp. AIRB-ML5.3 | + | + | + | + | - | + |
| Sugiyamaella sp. AIRB-ML6.4 | + | + | + | + | - | + |
| Candida sp. AIRB-ML7.1 | + | + | + | + | - | + |
| Spencermartinsiella sp. AIRB-ML9.2 | + | - | + | + | - | + |
| Sugiyamaella sp. AIRB-ML11.1 | + | + | + | + | - | + |
+ = Positive test; - = Negative test.
The following molecules were tested as a carbohydrates source: cellobiose, fructose, galactose, glucose, mannose, and xylose. The strains AIRB-ML2.1 (Rhodotorula sp.), AIRB-ML5.1 (Leucosporidium sp.), AIRB-ML1.1 (Candida sp.), AIRB-ML4.1 (Sugiyamaella sp.), AIRB-ML5. 3 (Sugiyamaella valdiviana), AIRB-ML6.4 (Sugiyamaella sp.), and AIRB-ML11.1 (Sugiyamaella valdiviana) were able to assimilate cellobiose, fructose, galactose, glucose, and xylose. The AIRB-ML2.1 (Rhodotorula sp.) and AIRB-ML4.2 (Cryptococcus sp.) strains were the only ones capable of metabolizing mannose. AIRB-NL4.2 (Cryptococcus sp.) and AIRB-ML9.2 (Spencermartinsiella sp.) do not assimilate galactose, but assimilated, cellobiose, fructose, glucose, and xylose.
A qualitative test was carried out to determine if the assimilation of the tested carbohydrates was done through fermentative processes (CO2 production). The results indicated that Cryptococcus sp. (AIRB-ML4.2) ferment glucose, galactose, xylose, and mannose. Rhodotorula sp. (AIRB-ML2.1) was able to ferment glucose, xylose, and mannose, and Leucosporidium sp. (AIRB-ML5.1) was only able to ferment xylose (Figure 3).
Figure 3 Fermentative assimilation assays of carbohydrates by different strains isolated from degradation wood “Palo podrido”. A = Cryptococcus sp. (A1 = Glucose; A2 = Galactose; A3 = Xylose; A4 = Mannose); B = Rhodotorula sp. (B1 = Mannose; B2 = Glucose; B3 = Xylose); C = Leucosporidium sp. (C1 = Xylose).
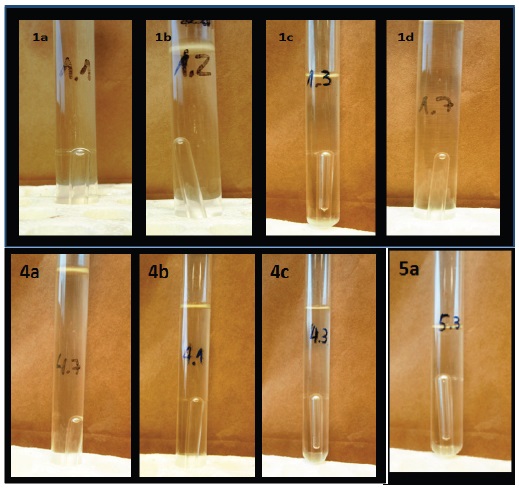
Figura 3. Pruebas de asimilación fermentativa de carbohidratos por las diferentes levaduras aisladas de la degradación de la madera “Palo podrido”. A = Cryptococcus sp. (A1 = Glucosa; A2 = Galactosa; A3 = Xilosa; A4 = Manosa); B = Rhodotorula sp. (B1 = Manosa; B2 = Glucosa; B3 = Xilosa); C = Leucosporidium sp. (C1 = Xilosa).
The results indicated that Cryptococcus sp. and Rhodotorula sp. presented the potential to produce ethanol using hexoses (glucose, galactose, and mannose) and pentoses (xylose). The case of Leucosporidium sp. showed fermentative metabolism only with xylose. The results suggest that the isolated microorganisms can assimilate simple carbon sources directing their metabolism towards biomass production and fermentative processes. The ability to metabolize simple sugars is consistent with the environment where they were isolated and may reflect adaptation to the environment.
Screening of strains with xylanolytic, cellulolytic, and laccase activity
Strains isolated from decomposing wood, “Palo podrido”, apart from assimilating the carbohydrates present in the degraded material, can also participate in the degradation of lignocellulose, and contribute to the mineralization of the wood. At this stage of the work, the ability to produce enzymes that can participate in lignocellulose degradation was studied. The study focused on identifying the enzymes cellulase, xylanase, and laccase. Lignin peroxidase nor manganese peroxidase enzyme activity was not detected. The results obtained are presented in Table 3. It is observed that only 5 out of the 10 isolated microorganisms presented cellulolytic activity: Rhodotorula sp. (AIRB-ML2.1), Cryptococcus sp. (AIRB-ML4.2), Leucosporidium sp. (ARIB-ML5.1), Candida sp. (ATRM-ML1.1), and Sugiyamaella sp. (ARIB-ML4.1). In comparison, 7 of the 10 isolated microorganisms presented xylanolytic activity, and these were Rhodotorula sp. (AIRB-ML2.1), Cryptococcus sp. (AIRB-ML4.2). Leucosporidium sp. (ARIB-ML5.1), Candida sp. (ATRM-ML1.1), Sugiyamaella sp. (ARIB-ML4.1), Sugiyamaella valdiviana (AIRB-ML6.4) and Candida chilensis (AIRB-ML7.1). Only the strain of Cryptoccocus sp. showed laccase activity.
Table 3. Determination of xylanolytic and cellulolytic activity of yeasts isolated from decomposing wood (Palo podrido) at 48 h of culture.
Tabla 3. Determinación de las actividades celulolítica y xilanolítica de las levaduras aisladas de la descomposición de madera (Palo podrido) a 48 h de cultivo.
| Clave | Assigned species | Xylanolytic activity (PI) | Celulolytic activity (PI) |
| AIRB-ML2.1 | Rhodotorula sp. | 1.4 ± 0.2a | 1.3 ± 0.2a,b |
| AIRB-ML4.2 | Cryptococcus sp. | 1.3 ± 0.0a | 1.3 ±0.0a,b |
| AIRB-ML5.1 | Leucosporidium sp. | 1.4 ± 0.0a | 1.2 ± 0.2a |
| AIRB-ML1.1 | Candida sp. | 1.2 ± 0.0a | 1.5 ± 0.1b |
| AIRB-ML4.1 | Sugiyamaella sp. | 1.2 ± 0.0a | 1.3 ± 0.1a,b |
| AIRB-ML5.3 | Sugiyamaella sp. | 0 ± 0.0b | 0 ± 0.0c |
| AIRB-ML6.4 | Sugiyamaella sp. | 1.3 ± 0.1a | 0 ± 0.0c |
| AIRB-ML7.1 | Candida sp. | 1.4 ± 0.0a | 0 ± 0.0c |
| AIRB-ML9.2 | Spencermartinsiella sp | 0 ± 0.0b | 0 ± 0.0c |
| AIRB-ML11.1 | Sugiyamaella sp. | 0 ± 0.0b | 0 ± 0.0c |
Data correspond to media ± SD using three observations. Different letters indicate significantly different (p < 0.05, Tukey-HSD); PI = Power Index.
Effect of nitrogen source on cellulolytic and xylanolytic activity
The strains that presented the highest hydrolytic enzyme activities (cellulase and xylanase) were selected for the next stage, in which the effect of the type and concentration of the Nitrogen source on the production of hydrolytic enzymes was studied. Five types of peptones of different origins and total Nitrogen content were used to verify the effect of nitrogen on the growth of microorganisms and enzyme production. Three total Nitrogen concentrations (1, 2, and 3 %) were tested, and CMC and birch xylan were maintained as carbon sources.
Rhodotorula sp. showed xylanolytic activity in the medium supplemented with CP-H in all tested concentrations (1, 2, and 3 % of total N) (Table 4). Significant differences were found in the xylanolytic activity of Rhodotorula sp. with 2 and 3 % of total nitrogen in the culture medium. Leucosporidium sp. presented xylanolytic activity in CP-H and PP-3 BD (Table 4).
Table 4. Effect of type and total nitrogen concentration on Rhodotorula sp. (AIRB-ML2.1) and Leucosporidium sp. (AIRB-ML5) xylanolytic activity.
Tabla 4. Efecto del tipo y concentración total de nitrógeno sobre la actividad xilanolítica de Rhodotorula sp. (AIRB-ML2.1) y Leucosporidium sp. (AIRB-ML5)
| Nitrogen type | Total Nitrogen (%) | Xylanolytic activity in plate (PI) | |||||
| Rhodotorula sp. AIRB-ML2.1 | Leucosporidium sp. AIRB-ML5.1 | ||||||
| Concentration (%) | Concentration (%) | ||||||
| 1 | 2 | 3 | 1 | 2 | 3 | ||
| CP-H | 7.481 ± 0.030 | 1.0 ± 0.0 | 1.4 ± 0.0a | 1.3 ± 0.1a | 1.0 ± 0.0 | 0 ± 0.0a | 0 ± 0.0 |
| CP-H2 | 8.774 ± 0.012 | 0 ± 0.0 | 0 ± 0.0b | 1.6 ± 0.1b | 0 ± 0.0 | 0 ± 0.0a | 0± 0.0 |
| PP-3 | 12.868 ± 0.093 | 0 ± 0.0 | 1.3 ± 0.1a | 1.4 ± 0.0a | 0 ± 0.0 | 1.0 ± 0.1b | 1.0 ± 0.0 |
| BT | 13.110 ± 0.022 | 0 ± 0.0 | 1.2 ± 0.2a | 1.0 ± 0.1c | 0 ± 0.0 | 0 ± 0.0a | 0 ± 0.0 |
| GP | 15.140 ± 0.027 | 0 ± 0.0 | 1.6 ± 0.1c | 0 ± 0.0d | 0 ± 0.0 | 0 ± 0.0a | 0 ± 0.0 |
Data correspond to media ± SD using three observations. The different letters indicated significantly different (p < 0.05, Tukey-HSD).
Regarding cellulolytic activity, it was observed that Rhodotorula sp. (AIRB-ML 2.1) showed plaque activity in culture media supplemented with CP-H (2 and 3 % of total nitrogen) and CP-H2 (1 and 2 % of total nitrogen) (Table 5). In comparison, Leucosporidium sp. (AIRB-ML5.1) showed cellulolytic activity in plates with CP-H (3 % of total nitrogen) and CP-H2 (2 % of total nitrogen).
Table 5. Effect of type and total N concentration on Rhodotorula sp. (AIRB-ML2.1) and Leucosporidium sp. (AIRB-ML5.1) cellulolytic activity.
Tabla 5. Color of squash fruit peel (Cucurbita pepo L.) var. ‘Grey Zucchini’s.
| Nitrogen type | Total Nitrogen (%) | Cellulolytic activity in plate (PI in cm) | |||||
| Rhodotorula sp. AIRB-ML2.1 | Leucosporidium sp. AIRB-ML5.1 | ||||||
| Concentration (%) | Concentration (%) | ||||||
| 1 | 2 | 3 | 1 | 2 | 3 | ||
| CP-H | 7.481 ± 0.030 | 0 ± 0.0a | 1.1 ± 0.1a | 1.2 ± 0.1a | 0 ± 0.0 | 0 ± 0.0 | 1.0 ± 0.0 |
| CP-H2 | 8.774 ± 0.012 | 1.8 ± 0.2b | 1.5 ± 0.0b | 0 ± 0.0b | 0 ± 0.0 | 0 ± 0.0 | 0.9 ± 0.0 |
| PP-3 | 12.868 ± 0.093 | 0 ± 0.0a | 0 ± 0.0c | 0 ± 0.0b | 0 ± 0.0 | 0 ± 0.0 | 0 ± 0.0 |
| BT | 13.110 ± 0.022 | 0 ± 0.0a | 0 ± 0.0c | 0 ± 0.0b | 0 ± 0.0 | 0 ± 0.0 | 0 ± 0.0 |
| GP | 15.140 ± 0.027 | 0 ± 0.0a | 0 ± 0.0c | 0 ± 0.0b | 0 ± 0.0 | 0 ± 0.0 | 0 ± 0.0 |
Efecto del tipo y concentración total de nitrógeno sobre la actividad celulolítica de Rhodotorula sp. (AIRB-ML2.1) y Leucosporidium sp. (AIRB-M-5.1)
Rhodotorula sp. produced xylanases in all media. The best results were obtained with the medium supplemented with CP-H (enzymatic digest of casein that does minor damage to casein vitamins and amino acids such as tryptophan) at 1, 2, and 3 %. The highest xylanolytic activity was observed in the medium supplemented with 2 % GP. (Pancreatic digest of pigskin with a low concentration of fermentable carbohydrates) and in the medium supplemented with 3 % GP-H2 (acid hydrolysate of casein that racemizes amino acids and destroys vitamins). This enzymatic activity was also observed in the media supplemented with PP-3 (enzymatic digest of porcine proteins with high protease content and variable concentrations of vitamins and minerals) at 2 and 3 % and in the medium supplemented with BT (pancreatic hydrolysate of casein with low carbohydrate levels) at 2 and 3 %.
Leucosporidium sp. only presented xylanolytic activity in the media supplemented with CP-H (1 %) and PP-3 (2 and 3 %); it can be deduced that the presence of vitamins is necessary for its growth; both Nitrogen sources provide vitamins in the growth medium. The xylanolytic activity was not observed in the medium supplemented with CP-H2, where the vitamins and amino acids were completely hydrolyzed. A similar result was observed in media supplemented with BT and GP, where pancreatic digestion of casein destroys the biological quality of vitamins and proteins, not allowing optimal growth.
Regarding the effect of the Nitrogen source on cellulolytic activity, Rhodotorula sp. produced extracellular cellulases in the media supplemented with CP-H (2 and 3 %) and CP-H2 (1 and 2 %); both media differ only in the form of the hydrolyzed casein.
Discussion
The molecular identification of the isolated strains was carried out based on analyzing the ITS region sequences widely used for these purposes (Takemoto et al., 2008). Each sequence was compared with the GenBank database deposited in the NCBI; for the identification of the strains, the following criteria were used: expectation value (> 0.01) and identity percentage (> 98 %) (Baffi et al., 2011).
The results obtained for the isolated strains are consistent with those reported by other authors, basidiomycetes of the genera Rhodotorula (Ramírez and González 1985), Cryptococcus (Mazza et al., 2013; Lara et al., 2014) and Leucosporidium of degraded wood such as the one studied in this work. Ascomycetes have also been isolated as Sugiyamaella (Morais et al., 2013; Lara et al., 2014), Candida (Cadete et al., 2012; Guo et al., 2012; Morais et al., 2013), and Spencermartinsiella (Morais et al., 2013) among others.
During the enzymatic hydrolysis of lignocellulosic material carried out by basidiomycete fungi, biopolymers such as hemicellulose and cellulose are hydrolyzed, releasing a complex mixture of monosaccharides and oligosaccharides (Zadrazil et al., 1982). Many non-ligninolytic microorganisms (bacteria and yeast) use the sugars derived from hydrolysis, being part of the biota associated with wood degradation natural processes. The assimilation of hexoses is the most common metabolic process in bacteria and yeasts, and to a minor degree, the assimilation of pentoses. Of the isolated yeasts, most were able to metabolize both hexoses and pentoses; only the AIRB-ML2.1 (Rhodotorula sp.) and AIRB-ML4.2 (Cryptococcus sp.) strains showed the ability to assimilate mannose (Table 2). Microorganisms such as Rhodotorula sp., Rhodotorula mucilaginiosa, Rhodotorula graminis, and Rhodotorula glutinis can assimilate glucose, mannose, sucrose, xylose, and mixtures of xylose, and glucose (Martins et al., 2018; Hamidi et al., 2020). On the other hand, it was reported that Candida sp., Candida melibiosica, Candida dubliniensis, and Candida albicans can metabolize maltose, trehalose, cellobiose, melibiose, raffinose, sucrose, lactose, glycerol, sorbitol, glucose, fructose, and xylose (Gientka et al., 2016). Similarly, the assimilation of glucose, xylose, and cellobiose in Cryptococcus sp. and Cryptococcus curvatus were described (Yu et al., 2014; Diamantopoulou et al., 2020). Sugiyamaella americana can assimilate arabinose, cellobiose, glucose, galactose, ribose, sorbose, trehalose, and xylose, among others (Kurtzman 2011). The “Palo podrido” degradation that some Ganoderma species carry out is a degradation in which an extracellular medium rich in monosaccharides, polysaccharides, and compounds derived from lignin is generated (Ho et al., 2020). The complete mineralization of this medium is completed by the opportunistic microbiota composed mainly of bacteria and yeasts (Tláskal et al., 2021). The results showed that the isolated microorganisms mainly metabolize the monosaccharides via assimilation (biomass generation).
Concerning the fermentative metabolism observed, the results obtained allow us to establish that Cryptococcus sp. was able to use galactose, xylose, and mannose via fermentation, contrary to that described by Chang et al. (2015) and Xu et al. (2014), who reported the use of xylose, galactose, and mannose to produce lipids. The ability of Cryptococcus sp. to ferment these sugars can be an advantage since fermentative processes could be generated to produce ethanol, being an advantage over yeasts such as Saccharomyces cerevisiae which is only capable of fermenting glucose. In the case of Rhodotorula sp., it showed the ability to ferment glucose, mannose, and xylose. Compared with other reports, it was observed that Rhodotorula glutinis and Rhodotorula paludigena were used to produce lipids from glucose and xylose (Gosalawit et al., 2021), while Bura et al. (2012), and Nadal et al. (2020) reported ethanol production using Rhodotorula mucilaginosa and R. glutinis with galactose, mannose, and a mixture of glucose-xylose as substrates.
The next microorganism that showed fermentative capacity was Leucosporidium sp. with xylose as a substrate. De mot et al. (1985) and Tasseli et al. (2018) reported that Leucosporidium capsuligenum and Leucosporidium cratinovorum produced ethanol using concentrated dextrin solutions (25 % v/v) and a glucose-xylose mixture as substrates. The fermentation of carbohydrates produces CO2 in parallel; one way to estimate this process is to observe the production of this gas using Durham hoods, a widely used qualitative method (Brooks, 2008; Wu et al., 2020). The results obtained in this stage of the work agree with those described by other authors. Most yeasts can assimilate glucose, fructose, and mannose (Flores et al., 2000) through the glycolytic pathway for biomass generation; however, under anaerobic conditions, pyruvate is oxidized to CO2 through the tricarboxylic acid cycle to generate ethanol. Yeasts can also metabolize pentoses through the pentose-phosphate pathway, which, depending on environmental conditions, can contribute to glucose catabolism through the trioses formed (Maaheimo et al., 2002). Cryptococcus sp., Rhodotorula sp., and Leucosporidium sp. can assimilate monosaccharides such as glucose, mannose, and xylose via fermentation of great biotechnological interest, making it necessary to generate adequate growth media to optimize ethanol production.
Regarding the hydrolytic activity studied, cellulase production has been observed in Rhodotorula sp., R. mucilaginosa, and Rhodotorula slofiae (Brandão et al., 2011; Rani et al., 2015). On the other hand, Carrasco et al. (2012), and Shariq and Sohail (2020) described cellulolytic activity in Candida gastricus, Candida stellate, Candida pullcherrima, and Candida tropicalis.Brandão et al. (2011) isolated and identified Cryptococcus adeliensis strains with cellulolytic activity, and Turkiewicz et al. (2005) described cellulolytic activity in Leucosporidium antarcticum.
In the case of xylanolytic activity, Duarte et al. (2013) described this activity in Rhodotorula strains, Lara et al. (2014), and Bastawde et al. (1994) described xylanolytic activity in Cryptococcus diffluens, Cryptococcus heveanensis, Cryptococcus laurentii, Cryptococcus albidus, and Cryptococcus flavus. Morais et al. (2020), and Shariq and Sohailm (2020) described xylanolytic activity in Candida pullcherrima, Candida colliculosa, Candida stellata, Candida oleophila, Candida valida, Candida hellenica, Candida pelliculosa, C. tropicalis, and Candida pseudolambica. Finally, Duarte et al. (2013) reported xylanolytic activity in Leucosporidium scottii. The hydrolytic enzymatic activity of isolated microorganisms in “Palo podrido” degradation may be associated with monosaccharides and oligosaccharides resulting from a primary degradation of lignocellulosic material of basidiomycetes fungi.
From the isolated strains, Cryptococcus sp. is the only strain that presented laccase activity. Laccase in Cryptococcus has been related to infective processes in humans (Sidrim et al., 2013). This enzyme was also isolated from a strain of Cryptococcus albidus that grew in sediments of paper pulp (Singhal et al., 2021). We observed that glucose and xylose were catabolic repressors of laccase at concentrations of 0.5 % or more. As described by Chen and Williamson (2011), in Cryptococcus neoformans, glucose is a repressor of laccase expression; these results also coincide with those reported by Zhu and Williamson (2004) and Lazera et al. (1996), who observed that glucose acts as a metabolic regulator since the low concentration of glucose stimulates the expression of laccase, improving the delignification process in decaying wood (Lazera et al., 1996). The laccase activity detected in Cryptococcus sp. can contribute to the degradation of compounds of phenolic origin resulting from the hydrolysis of wood lignin. This enzymatic activity possibly contributes to reducing the toxicity of the growth medium, generating better conditions for other microorganisms sensitive to the presence of phenolic compounds.
The nitrogen naturally present in lignocellulolytic residues is very low (Martius 1992), so basidiomycete fungi have evolved to grow in environments where the C/N ratio is very high. Regarding the Nitrogen sources tested, for both strains evaluated, the variation in the percentage of peptone in the culture medium did not directly correlate with the xylanolytic activity observed. Studies of xylanase-producing in fungi have focused on studying the effect of the carbon source and the incubation temperature on enzymatic activity (Lara et al., 2014). The relationship between nitrogen source and enzyme activity is not yet evident in basidiomycete fungi and some yeasts. Haapala et al. (1996) determined the effect of the Nitrogen source on xylanolytic activity and endo -1,4 - β-glucanase, establishing that the culture medium supplemented with protease peptone and yeast extract favors the production of both enzymes. In Aspergillus niger, peptone was the best Nitrogen source compared to yeast extract, (NH4)2SO4, and urea to promote xylanase activity (Betini et al., 2009).
In filamentous fungi, modification of the culture medium is a strategy to improve cellulase production; in an investigation carried out with Aspergillus terreus, Shahriarinour et al. (2011) studied the effect of the Nitrogen source at different concentrations on cellulolytic activity and observed that yeast extract promotes the formation of the cellulolytic complex (FPase, CMCase, β-glucosidase) while peptone was better than urea and (NH4)2SO4. Gomes et al. (1989) evaluated the effect of the type of peptone (meat peptone, bacto peptone, and soy peptone), cellulosic substrates, inorganic nitrogen, and urea on the production of cellulases of Gliocladium virens. They found that cellulose and peptone in the medium are necessary to produce enzymes. Similarly, a work by Liang et al. (2010) with Anoxybacillus sp. determined that peptone as a Nitrogen source in the culture medium registered the highest cellulolytic activity to yeast extract, tryptone, or inorganic Nitrogen sources.
By analyzing the compositions of each Nitrogen source, Rhodotorula sp. may need essential amino acids and vitamins with minimal degradation for optimal growth. The results obtained with the medium supplemented with CP-H2 at 3 % corroborate these observations; this Nitrogen source provides degraded amino acids and vitamins. Under this condition, xylanolytic activity is observed. The need for Nitrogen and vitamins of Rhodotorula sp. at this concentration is probably covered by reactivating enzyme production.
An excess in the Nitrogen concentration can generate problems in the expression of hydrolytic enzymes (Edwards et al., 2011); this result differs from what was found by Elisashvili et al. (2011), who observed that the addition of nitrogen accelerates the growth of fungi, increasing the secretion of hydrolytic enzymes associated with growth. The differences observed are due to the origin of the Nitrogen present in the culture medium; basidiomycete fungi respond better to organic Nitrogen sources such as peptone and yeast extract, which affect microbial growth, while inorganic Nitrogen sources can affect metabolism by the lack of growth factors necessary for the development of the fungus. The addition of nitrogen affected the secretion of hydrolytic enzymes (cellulases and xylanases) in Leucosporidium sp., while in Rhodotorula sp., it only affected the secretion of xylanase. The metabolism of nitrogenous compounds is very complex. In basidiomycete, fungus is mediated by extracellular enzymes such as alanine transaminase, urease, aspartate transaminase, aspartase, glutamine synthetase, glutamate dehydrogenase, and nitrate reductase. These enzymes hydrolyze proteins into amino acids that are subsequently introduced into the cell with the help of permeases (Grotjohann et al., 2014). Therefore, it is expected that both the degree of hydrolysis and the quality of the proteins (amino acids content) used to complement the growth media are essential to increase the growth of the microorganisms under study.
Conclusions
In the present work, yeasts of the phylum Basidiomycota of the genera Rhodotorula, Cryptococcus, Leucosporidium, and the phylum Ascomycota of the genera Candida, Sugiyamaella, and Spencermartinsiella were isolated and identified from the degradative process of wood known as “Palo podrido” present in the Ulmo tree (Euryphia cordifolia). The lignocellulolytic capacity of the isolated strains was studied, and Rhodotorula sp. (AIRB-ML2.1) and Leucosporidium sp. (AIRB-ML5.1) were able to produce extracellular cellulases and xylanases. None of the ten isolated strains grew on media supplemented with a complex carbon source (potato starch). Only Cryptococcus sp. produced laccase activity with glycerol as a carbon source in the culture medium or concentrations less than 0.5 % of glucose. Cryptococcus sp., Leucosporidium sp., and Rhodotorula sp. showed fermentative metabolism. The origin and concentration of Nitrogen sources could influence the production of extracellular hydrolytic enzymes. The isolated yeasts showed great metabolic diversity, highlighting their ability to assimilate monosaccharides and the production of hydrolytic enzymes. This makes them of interest for their possible use in biotechnological processes.











 nueva página del texto (beta)
nueva página del texto (beta)




