Introduction
The biological activity of organotin(IV) compounds and their use as metallopharmaceuticals has been reported [1-13]. A tremendous research is directed towards the design of non-platinum chemotherapeutics with the aim to optimize the features of classical platinum drugs constituting the basic cisplatin framework viz. their toxic side effects, inherent intrinsic resistance and high cost [14]. Diorganotin(IV) complexes are acting as potential antitumours. They exhibit promise activity against many tumour cell lines [15-19]. Numerous diorganotin(IV) derivatives have been found to exhibit high in vivo cytotoxicity against P388 lymphocytic leukaemia.
The antitumour action mechanism is still unclear, although some evidence is consistent with their binding directly to DNA. The antituour activity of the organotin(IV) complex R2SnX2L is depending on the group attached to the tin atom, leaving groups (X) and the coordinated ligand (L). The coordinated ligand favours in some way the transport of the drug into cells, and the antitumour activity would be exerted by the diorganotin(IV) ion released from the complex [20]. The released diorganotin(IV) species would interact with nucleic acids, similarly as in the case of the widely used anticancer drug cisplatin. Therefore, there is a relationship between the stability of the organotin(IV) complexes and their antitumour activity. In conjunction with our previous studies on organotin(IV) complexes [21-26] the present paper aims to study the dimethyltin(IV) complexes with N,N,N’,N’-tetraethylethylene diamine to have information regarding the plausible interaction mechanism toward DNA fragment. The four ethyl groups attached to ethylenediamine nitrogen atoms may undergo hydrophobic interaction with the methyl groups of tin(IV) moiety. Such interaction may increase the stability of the formed complex. Also, the four ethyl groups of Et4en will increase the hydrophobic nature of the complex, which will facilitate the dimethyltin(IV) complex to get into the cell. Density functional theory (DFT) calculations for geometry optimization and vibrational frequencies of all complexes and ligands were reported.
Experimental
Materials and Reagents
Dimethyltin(IV) dichloride (DMT) was obtained from Merck Chem.Co. The ligand N,N,N’,N’-tetraethylethylenediamine was supplied by Sigma Chem. Co., 1,4-Dioxane was provided by Aldrich Chem. Co. Sodium hydroxide solutions were prepared by diluting the content of BDH concentrated volumetric solutions vials. NaOH solutions were frequently checked by titration against potassium hydrogen phthalate.
Procedure and Measuring Techniques
A Metrohm 686 titroprocessor was used for the potentiometric titrations. The titroprocessor is equipped with a 665 dosimat (Switzerland-Herisaue) and a Metrohm glass-calomel combined electrode is used. NBS standard buffer solutions [27] were used to calibrate the titroprocessor and electrode. The temperature of the titrated solutions was adjusted by circulating thermostatically controlled water through the jacket of a titration vessel. Sodium nitrate was used to adjust the ionic strength to 0.1 mol dm-3. All titrations were performed under a nitrogen atmosphere. N,N,N’,N’-tetraethylethylenediamine solution was prepared in the protonated form by dissolving in HNO3 solution. The protonation constants of Et4en were determined by titrating 1.25 mmol. The hydrolysis constants of dimethyltin(IV) were determined by titrating 1.25 mmol. The formation constants of Et4en complexes with dimethyltin(IV) were determined by titrating solution mixture of Et4en (1.25 mmol) and dimethyltin(IV) (1.25 mmol). The volume of each titrated solution mixtures is reached to 40 mL and the ionic strength was 0.1 mol. dm-3 (adjusted with NaNO3). 0.05 mol. dm-3 NaOH solution was used as titrant. The pKw values of solution mixtures of dioxane-H2O were estimated as reported previously [28,29]. In this procedure various amounts of standard NaOH solution were added to a 0.10 mol. dm-3 NaNO3 solution. From the amount of base added, [OH-] was calculated. [H+] was calculated from the pH value. The values obtained in this way for log[OH-][H+] (log Kw) are -14.23, -14.50, -14.92, -15.12 and -15.63 for 25.0, 37.5, 50.0, 62.5 and 75.0% dioxane in H2O, respectively.
The titration data were processed to evaluate the equilibrium constants expressed by Eq. 1 and Eq. 2.
Where M, L and H stand for dimethyltin(IV), Et4en and proton respectively. The compound of general formula MpLqHr formed is expressed by the coefficients p, q and r, where M, L and H stand for dimethyltin(IV), Et4en and proton respectively. The formation constants were evaluated using the computer program MINIQUAD-75 [30]. The stoichiometry and formation constants of the complexes formed were determined by trying various possible models of different composition. The accepted model is giving the best statistical fit and chemically consistent with the magnitude of various residuals, as reported previously [30]. Tables 1 and 2 include the formation constants and their standard deviation as obtained from the MINIQUAD output. The speciation diagrams were evaluated using the program SPECIES [31].
Table 1 Formation constants of dimethyltin (IV) complexes with Et4en in water at different temperatures.
| Systema | Temp(°C) | p | q | rb | Logβc | Sd |
|---|---|---|---|---|---|---|
| DMT | 15 | 1 | 0 | -1 | -3.56(0.01) | 5.4E-8 |
| 1 | 0 | -2 | -9.05(0.01) | |||
| 1 | 0 | -3 | -19.79(0.04) | |||
| 1 | 0 | -4 | -30.41(0.04) | |||
| 2 | 0 | -2 | -4.23(0.01) | |||
| 2 | 0 | -3 | -9.52(0.01) | |||
| 2 | 0 | -4 | -15.23(0.01) | |||
| DMT-Et4en | 15 | 0 | 1 | 1 | 9.94(0.01) | 1.5E-8 |
| 0 | 1 | 2 | 16.80(0.01) | |||
| 1 | 1 | 0 | 11.71(0.03) | 8.9E-10 | ||
| 1 | 2 | 0 | 18.53(0.06) | |||
| 1 | 1 | 1 | 16.69(0.05) | |||
| 1 | 1 | -1 | 5.32(0.05) | |||
| 1 | 1 | -2 | -2.52(0.05) | |||
| DMT | 20 | 1 | 0 | -1 | -3.31(0.01) | 6.1E-8 |
| 1 | 0 | -2 | -8.64(0.01) | |||
| 1 | 0 | -3 | -19.27(0.05) | |||
| 1 | 0 | -4 | -30.71(0.02) | |||
| 2 | 0 | -2 | -3.77(0.01) | |||
| 2 | 0 | -3 | -8.91(0.02) | |||
| 2 | 0 | -4 | -14.44(0.01) | |||
| DMT-Et4en | 20 | 0 | 1 | 1 | 9.81(0.01) | 1.4E-8 |
| 0 | 1 | 2 | 16.54(0.01) | |||
| 1 | 1 | 0 | 11.52(0.04) | 3.2E-8 | ||
| 1 | 2 | 0 | 18.23(0.05) | |||
| 1 | 1 | 1 | 16.47(0.05) | |||
| 1 | 1 | -1 | 5.17(0.04) | |||
| 1 | 1 | -2 | -2.41(0.04) | |||
| DMT | 25 | 1 | 0 | -1 | -3.03(0.01) | 4.3E-8 |
| 1 | 0 | -2 | -8.21(0.01) | |||
| 1 | 0 | -3 | -18.73(0.03) | |||
| 1 | 0 | -4 | -29.54(0.02) | |||
| 2 | 0 | -2 | -3.12(0.01) | |||
| 2 | 0 | -3 | -8.13(0.02) | |||
| 2 | 0 | -4 | -13.59(0.02) | |||
| DMT-Et4en | 25 | 0 | 1 | 1 | 9.65(0.01) | 3.4E-8 |
| 0 | 1 | 2 | 16.22(0.02) | |||
| 1 | 1 | 0 | 11.31(0.02) | 4.1E-10 | ||
| 1 | 2 | 0 | 17.80(0.04) | |||
| 1 | 1 | 1 | 16.23(0.04) | |||
| 1 | 1 | -1 | 5.00(0.04) | |||
| 1 | 1 | -2 | -2.24(0.05) | |||
| DMT | 30 | 1 | 0 | -1 | -2.81(0.01) | 6.6E-8 |
| 1 | 0 | -2 | -7.91(0.02) | |||
| 1 | 0 | -3 | -18.38(0.05) | |||
| 1 | 0 | -4 | -28.71(0.02) | |||
| 2 | 0 | -2 | -2.87(0.02) | |||
| 2 | 0 | -3 | -7.84(0.04) | |||
| 2 | 0 | -4 | -13.06(0.04) | |||
| DMT-Et4en | 30 | 0 | 1 | 1 | 9.51(0.01) | 9.3E-9 |
| 0 | 1 | 2 | 15.94(0.01) | |||
| 1 | 1 | 0 | 11.08(0.03) | 8.7E-10 | ||
| 1 | 2 | 0 | 17.32(0.09) | |||
| 1 | 1 | 1 | 15.98(0.04) | |||
| 1 | 1 | -1 | 4.81(0.07) | |||
| 1 | 1 | -2 | -2.11(0.05) | |||
| DMT | 35 | 1 | 0 | -2.49(0.02) | 6.9E-8 | |
| 1 | 0 | -2 | -7.54(0.02) | |||
| 1 | 0 | -3 | -17.95(0.06) | |||
| 1 | 0 | -4 | -28.20(0.03) | |||
| 2 | 0 | -2 | -2.27(0.03) | |||
| 2 | 0 | -3 | -7.11(0.04) | |||
| 2 | 0 | -4 | -12.34(0.04) | |||
| DMT-Et4en | 35 | 0 | 1 | 1 | 9.42(0.01) | 1.7E-8 |
| 0 | 1 | 2 | 15.70(0.01) | |||
| 1 | 1 | 0 | 10.90(0.05) | 1.1E-9 | ||
| 1 | 2 | 0 | 16.96(0.10) | |||
| 1 | 1 | 1 | 15.78(0.05) | |||
| 1 | 1 | -1 | 4.66(0.07) | |||
| 1 | 1 | -2 | 2.90(0.05) |
a p, q and r are the stoichiometric coefficient corresponding to organotin(IV), Et4en and H+, respectively; c Standard deviations are given in parentheses; d Sum of square of residuals.
Table 2 Formation constants of dimethyltin (IV) complexes with Et4en in dioxane-water solutions of different compositions.
| System | Dioxane % | p | q | ra | Logβb | Sc |
|---|---|---|---|---|---|---|
| DMT | 12.5 | 1 | 0 | -1 | -3.27(0.01) | 1.3E-07 |
| 1 | 0 | -2 | -8.70(0.02) | |||
| 1 | 0 | -3 | -19.53(0.06) | |||
| 1 | 0 | -4 | -30.09(0.03) | |||
| 2 | 0 | -2 | -3.68(0.02) | |||
| 2 | 0 | -3 | -8.94(0.05) | |||
| 2 | 0 | -4 | -14.52(0.04) | |||
| DMT-Et4en | 12.5 | 0 | 1 | 1 | 9.75(0.01) | 1.8E-08 |
| 0 | 1 | 2 | 16.40(0.02) | |||
| 1 | 1 | 0 | 11.37(0.02) | 1.5E-09 | ||
| 1 | 2 | 0 | 18.52(0.07) | |||
| 1 | 1 | 1 | 16.00(0.04) | |||
| 1 | 1 | -1 | 5.00(0.07) | |||
| 1 | 1 | -2 | -2.14(0.05) | |||
| DMT | 25 | 1 | 0 | -1 | -3.39(0.00) | 1.00E-08 |
| 1 | 0 | -2 | -8.99(0.01) | |||
| 1 | 0 | -3 | -19.83(0.01) | |||
| 1 | 0 | -4 | -31.30(0.01) | |||
| 2 | 0 | -2 | -3.92(0.01) | |||
| 2 | 0 | -3 | -9.27(0.04) | |||
| 2 | 0 | -4 | -15.11(0.03) | |||
| DMT-Et4en | 25 | 0 | 1 | 1 | 9.57(0.00) | 2.80E-09 |
| 0 | 1 | 2 | 16.02(0.01) | |||
| 1 | 1 | 0 | 11.11(0.03) | 1.50E-09 | ||
| 1 | 2 | 0 | 18.10(0.07) | |||
| 1 | 1 | 1 | 15.80(0.04) | |||
| 1 | 1 | -1 | 4.84(0.10) | |||
| 1 | 1 | -2 | -2.31(0.06) | |||
| DMT | 37.5 | 1 | 0 | -1 | -3.53(0.01) | 1.40E-08 |
| 1 | 0 | -2 | -9.23(0.01) | |||
| 1 | 0 | -3 | -20.18(0.02) | |||
| 1 | 0 | -4 | -31.83(0.01) | |||
| 2 | 0 | -2 | -4.17(0.01) | |||
| 2 | 0 | -3 | -9.61(0.03) | |||
| 2 | 0 | -4 | -15.59(0.03) | |||
| DMT-Et4en | 37.5 | 0 | 1 | 1 | 9.38(0.01) | 1.10E-08 |
| 0 | 1 | 2 | 15.72(0.01) | |||
| 1 | 1 | 0 | 10.72(0.02) | 1.30E-09 | ||
| 1 | 2 | 0 | 17.50(0.02) | |||
| 1 | 1 | 1 | 15.48(0.04) | |||
| 1 | 1 | -1 | 4.53(0.01) | |||
| 1 | 1 | -2 | -2.64(0.04) | |||
| DMT | 50 | 1 | 0 | -1 | -3.60(0.01) | 2.80E-08 |
| 1 | 0 | -2 | -9.43(0.01) | |||
| 1 | 0 | -3 | -20.58(0.02) | |||
| 1 | 0 | -4 | -32.52(0.02) | |||
| 2 | 0 | -2 | -4.30(0.01) | |||
| 2 | 0 | -3 | -9.78(0.04) | |||
| 2 | 0 | -4 | -16.03(0.04) | |||
| DMT-Et4en | 50 | 0 | 1 | 1 | 9.17(0.01) | 1.50E-08 |
| 0 | 1 | 2 | 15.30(0.01) | |||
| 1 | 1 | 0 | 10.31(0.04) | 7.50E-10 | ||
| 1 | 2 | 0 | 16.90(0.07) | |||
| 1 | 1 | 1 | 15.13(0.06) | |||
| 1 | 1 | -1 | 4.25(0.09) | |||
| 1 | 1 | -2 | -2.94(0.06) | |||
| DMT | 62.5 | 1 | 0 | -1 | -3.68(0.01) | 8.70E-08 |
| 1 | 0 | -2 | -9.62(0.02) | |||
| 1 | 0 | -3 | -20.89(0.04) | |||
| 1 | 0 | -4 | -33.26(0.05) | |||
| 2 | 0 | -2 | -4.47(0.01) | |||
| 2 | 0 | -3 | -9.90(0.03) | |||
| 2 | 0 | -4 | -16.51(0.04) | |||
| DMT-Et4en | 62.5 | 0 | 1 | 1 | 9.04(0.01) | 1.10E-08 |
| 0 | 1 | 2 | 15.11(0.01) | |||
| 1 | 1 | 0 | 9.93(0.03) | 7.40E-10 | ||
| 1 | 2 | 0 | 16.29(0.09) | |||
| 1 | 1 | 1 | 14.82(0.05) | |||
| 1 | 1 | -1 | 3.96(0.07) | |||
| 1 | 1 | -2 | -3.26(0.06) |
ap, q and r are the stoichiometric coefficient corresponding to organotin(IV), Et4en and H+ respectively; bStandard deviations are given in parentheses; cSum of square of residuals.
Computational Details
For geometries optimization, the density functional theory was applied using the Gaussian09 program [32]. A full optimization of all complexes and ligands geometries were obtained using the hybrid functional B3LYP (Becke's, three-parameter exchange functional, in combination with Lee-Yang-Parr correlation function) [33-35]. The basis set used is LANL2DZ (Los Alamos National Laboratory 2 double-zeta) [36-38] for Sn metal, while C, H, N, O and Cl were described by 6-311G* standard basis set in optimization. The GaussView 5.0 by Gaussian Inc. program was used for inspecting the input and output files generated by Gaussian09, for preprocessing, structure modification, and post processing analyses of structures, frequencies, and forces. To identify the most stable structure, the minima, a frequency analysis was performed for each stationary point. These analyses are performed to ensure that all minima have no imaginary frequencies in the vibrational mode calculations.
Results and discussion
Dimethyltin(IV) complex formation equilibria
The protonation constants of N,N,N’,N’-tetraethylethylenediamine were determined using the same experimental conditions as solvent composition, temperature and ionic strength which are used for the investigation of organotin complex equilibria. The overall protonation constants (logβ11 and logβ12) for Et4en were estimated. The values obtained agree with the literature data [39] considering changes in experimental condition.
The acid-base equilibria of dimethyltin(IV) ion was investigated previously in aqueous media [40-44]. The potentiometric data in the mixed solvents used were fitted assuming a model composed of the species 10-1, 10-2, 10-3, 10-4, 10-4, 20-3 and 20-4, Table 1. The aqua-hydroxo species (10-1) may undergo dimerization according to the general equilibrium
The dimerization constant (Kd) can be determined by Eq. 4.
Following the same way the dimerization constant of the species 20-4 is determined by the Eq. 5 [24].
The potentioetric titration curves of the protonated N,N,N’,N’-tetraethylethylenediamine and its dimethyltin(IV) complex are compared. The complex titration curve is lower than the Et4en curve, Fig. 1. This corresponds to the formation of a complex species through the ionization of hydrogen ion. The titration data were fitted considering the formation of the 110, 111, 11-1 and 11-2 species, according to scheme 1.
Eq. 6 and 7 [26] are used to calculate the pKa of coordinated water molecules.
The obtained values are 5.57 and 9.68 for pKa1 and pka2 respectively. These are higher than those of water molecules coordinated to the free dimethyltin(IV) ion. This may be explained on the premise that coordination of Et4en will decrease the electrophilicity of tin. This will result in the coordinated water molecule will be weakly bound and consequently will be less acidic.
The speciation diagram of DMT-Et4en complexes is used to provide a useful picture of the coordination of organotin with Et4en as a function of pH, Fig. 1. The protonated complex with stoichiometric coefficients 111, predominates at lower pH. It attains a maximum concentration of 38% at pH = 3.8. The deprotonated complex 110 starts to form at pH 3.0 and reaches the maximum concentration of 39% at pH = 5.7. The hydrolyzed form 11-1 predominates between pH = 5.7 and 8.0 with maximum formation degree of 27% at pH = 6.9. It is interesting to note that the complex species 110 and 11-1 are predominating in the physiological pH range. Therefore the binding of DMT complex with DNA constituent, the main target in the chemotherapy is possible. This may be explained on the premise that the DNA constituents coordinate in the negatively charged anionic form.
Effect of temperature
The thermodynamic parameters ΔH° and ΔS° associated with the protonation of N,N,N`,N`-tetraethylethylendiamine (Et4en) and its complex formation with the dimethyltin(IV) species were calculated from the temperature dependence of the data in Table 3. The plots are given in figures 2-3. These data can be employed to extrapolate the equilibrium constants to other temperatures. The main conclusions from the data can be summarized as follows:
The protonation reactions (8) and (10) of Et4en are exothermic and of comparable ΔH° value. Three factors affect the protonation reactions (i). The neutralization reaction, which is an exothermic process. (ii). Desolvation of ions, which is an endothermic process. (iii). The change of the conformation and the arrangements of the hydrogen bonds around the free and the protonated ligands.
The complexation reaction (11-15) for dimethyltin(IV) ion with Et4en is exothermic with negative (H˚ values, amounting to -70.0, -137.5, -78.5 and -57.1 kJ.mol-1.
Table 3 Thermodynamic parameters for the equilibria of dimethyltin(IV) complexes.a
| Equilibrium | ΔH° (kJmol-1) | ΔS° (JK-1 mol-1) |
|---|---|---|
| DMTb | ||
| 1) M(H2O)4 2++OH- ⇌ M(H2O)3(OH)++H2O | 40.2(0.5) | 344(2) |
| 2) M(H2O)3(OH)+ + OH- ⇌ M(H2O)2(OH)2+H2O | -11.5(0.2) | 128(1) |
| 3) M(H2O)2(OH)2 + OH- ⇌ M(H2O)(OH)3 -+H2O | -21.4(0.4) | -7(1) |
| 4) M(H2O)(OH)3 - + OH- ⇌ M(OH)4 2- + H2O | -20.9(0.4) | -4(1) |
| 5) 2M(H2O)4 2+ + 2OH- ⇌ M2(H2O)4(OH)2 2+ + 4H2O | 65.0(0.8) | 690(7) |
| 6) M2(H2O)4(OH)2 2++OH- ⇌ M2(H2O)3(OH)3 ++H2O | -13.0(0.3) | 127(1) |
| 7) M2(H2O)3(OH)3 + + OH- ⇌ M2(H2O)2(OH)4+H2O | -6.1(0.1) | 142(1) |
| Et4en | ||
| 8) L + H+ ⇌ LH+ | -45.6(0.1) | 32.0(0.3) |
| 9) LH+ + H+ ⇌ LH2 2+ | -45.78 | 31.98 |
| 10) L + 2H+ ⇌ LH2 2+ | -95.2(0.1) | -8.6(0.4) |
| DMT-Et4en | ||
| 11) M + L ⇌ ML | -70.0(0.1) | -18.6(0.3) |
| 12) M + 2L ⇌ ML2 | -137.5(0.3) | -121.4(1) |
| 13) M + L+ H ⇌ MLH | -78.5(0.1) | 47.1(0.3) |
| 14) M + L-H ⇌ MLH-1 | -57.1(0.1) | -96.0(0.3) |
| 15) M + L-2H ⇌ MLH-2 | 39.5(0.2) | 89.2(0.8) |
aM denotes (CH3)Sn2+; L denote; Standard deviations are given in parentheses. b hydrolysis data are from reference [51]
Effect of solvent
It is reported that the effective dielectric constant of the surrounding of active sites cavities of enzymes and proteins [45,46] is smaller , compared to in bulk water. Dielectric constants in such sites were found to be in range from 30 to 70 [47, 48]. Hence by using solution media containing around 10 to 50% dioxane, one may expect to approach to some extent the situation in these biological sites. By this way one may extrapolate the data to physiological condition. Careful examination of the medium effect on the equilibrium constants reveal the following features:
pKa of Et4en decrease linearly with increasing the percentage of organic solvent in the medium. This behavior is discussed based on the hydrophobic interaction between the neutral Et4en species and dioxane.
The pKa values of coordinated water molecules in [(CH3)2Sn(OH2)4] 2+ increase with increasing the dioxane percentage in the medium. This may be explained on the premise that the solvent of relatively low dielectric constant will increase the electrostatic forces between proton and hydrolyzed form of DMT.
The formation constants for dimethyltin(IV) complexes with Et4en decrease upon addition of dioxane to an aqueous solution of the corresponding species, figures 4-5. This can be explained by better solvation of virtually hydrophobic species CH3Sn+/CH3SnCl by dioxane through formation of coordinative bonds between O-dioxane and Sn centres. This leads to stabilization and modulation of solubility of dimethyltin(IV) species. This results in lowering the complex stability. This finding is in agreement with that proposed for alkyltin(IV) complexes with D-glucosamine [49].
Displacement reaction of coordinated Et4en by DNA constituents
It was reported that (CH3)2Sn(IV) has high affinity for N-donor ligands such as DNA constituents. Such interaction with DNA is thought to be responsible for the anti-tumour activity of related complexes [50]. The mechanism of antitumor activity of (CH3)2Sn-(amine) complex is based on the displacement of the amine by DNA. Consequently, the equilibrium constant for such reaction is of biological importance. Consider inosine as a typical DNA constituent (presented by HA) and Et4en (presented by B). The equilibria of complex-formation and displacement reactions may be presented introductory as:
The equilibrium constant for the displacement reaction is expressed by:
Substitution results in:
The formation constant value for Et4en complex, [(CH3)2SnB]2+ , amounts to log β110 = 11.31 at 25oC (Table 1). The log β110 values [51] for (CH3)2Sn-DNA constituents complexes [(CH3)2SnA]+ together with the equilibrium constants values (log Keq ) of displacement reaction calculated by Eq. 12, are given in Table 4. The Keq values clearly indicate the ability of DNA to displace the coordinated Et4en from its dimethyltin(IV) complex. Also, Keq value reveals to what extent the organotin-amine complex could interact with the DNA constituents, the main target in chemotherapy of tumours. The values of equilibrium constant of the displacement reaction for the different DNA constituents are compared. The nucleotide IMP and GMP have the highest values. The extra stability is due to the different columbic force of attraction between the dipositively charged dimethyltin(IV) ion and the nucleotides IMP or GMP having extra negative charges on the phosphate group. Also, the phosphate group of IMP and GMP is acting as anchoring site for tin [52]. This will facilitate the release of coordinated Et4en.
Table 4 Equilibrium constants for displacement reaction of coordinated N,N,N’,N’-tetramethylethylenediamine by DNA constituents at 25ºC and 0.1M NaNO3.
| DNA | logβ110 a DMT-DNA | logKeq |
|---|---|---|
| Inosine | 8.13 | -3.18 |
| Inosine-5΄-monophosphate | 11.90 | 0.59 |
| Guanosine-5΄- monophosphate | 12.34 | 1.03 |
| Adenine | 10.01 | -1.30 |
| Adenosine | 4.41 | -6.90 |
| Adenosine -5΄-monophosphate | 6.07 | -5.24 |
| Uracil | 9.34 | -1.97 |
| Thymine | 9.61 | -1.70 |
| Thymidine | 9.25 | -2.06 |
| Cytosine | 4.44 | -6.87 |
| Cytidine | 3.77 | -7.54 |
ataken from ref. [51]
Geometry optimiation
Calculations of the equilibrium geometries of the complexes and the ligands were carried out by the program Gaussian 09 at the B3LYP level of theory using the bases set LANL2DZ for Sn metal and standard basis set 6-311G* for C, H, N, O and Cl atoms. The optimized structures of the ligands and the complexes are given in Fig. 6. The geometric computational parameters for the optimized molecular geometry as bond angles, atomic charges and bond lengths are determined and given in (Tables 5 and 6). The results are interpreted as given in the following:
The N-CH2 bond length in the complexes ((CH3)2Sn(Et4en)Cl2, (CH3)2Sn(en)(H2O)2 2+, (CH3)2Sn(Et4en)(H2O)2 2+ and (CH3)2Sn(Et4en)(OH)2) are 1.50, 1.52 and 1.50 Å respectively. It is longer than the N-CH2 bond in the free ligand (1.47 Å). This elongation is due to the bonding of the Nitrogen atom with the metal.
The Sn-N bond length in the (CH3)2Sn(Et4en)Cl2 complex (2.53-2.54 Å) is longer than that in (CH3)Sn(en)Cl2 complex (2.40 Å). The main reason of this attributed to the changing the charge density on the nitrogen atom (-0.59 on N in en compared to -0.10 on N in Et4en).
The bond angle Et2N-C-C in Et4en is decreased from (115.8 °) upon complexation to 113.4 ° ((CH3)2Sn(Et4en)Cl2), 111.8 ° ((CH3)2Sn(Et4en)(H2O)2 2+), 113.6 ° ((CH3)2Sn(Et4en)(OH)2). This may be due to the formation of cyclic compounds.
The H2O-Sn(IV) bond length is 2.43-2.47 Å in (CH3)2Sn(Et4en)(H2O)2 2+ species while 2.39 Å in (CH3)2Sn(en)(H2O)2 2+ species. After deprotonation of coordinated water, the O-Sn(IV) bond is decreased to 2.00 Å for the two species. This is attributed to the strong bond between the negatively charged OH- ion and Sn(IV).
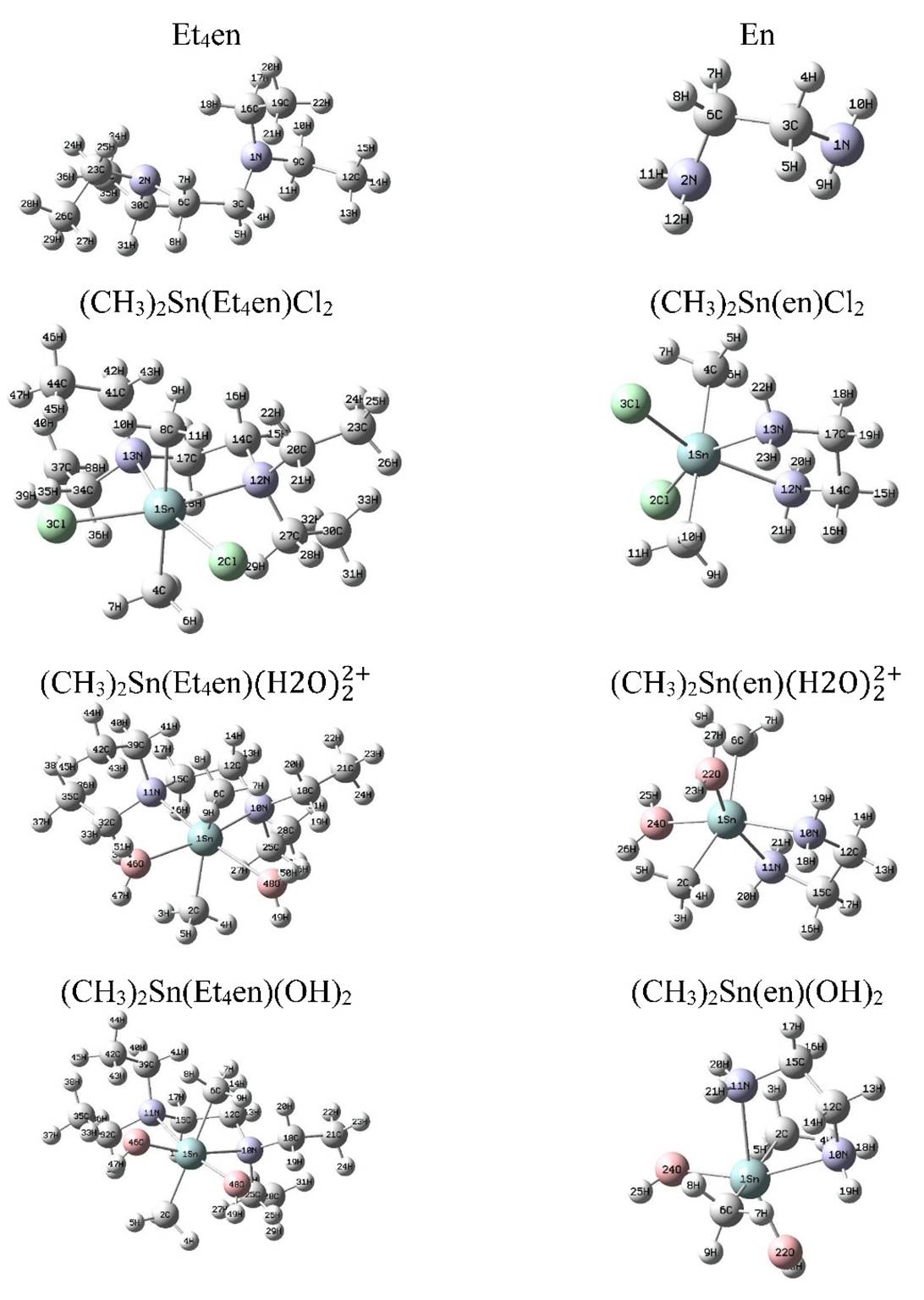
Fig. 6 The optimized structure of ligands and complexes according to B3LYP level of theory using the bases set LANL2DZ for Sn metal and standard basis set 6-311G* for C, H, N, O and Cl atoms.
Table 5 Calculated N−C, Sn−N and Sn−O bond lengths of the optimized structures of the ligands and complexes according to B3LYP/LanL2DZ level.
| Spices | d (N−C), Å | d (Sn−N), Å | d (Sn−O), Å | |||
|---|---|---|---|---|---|---|
| Et4en | N(2)-C(6) | 1.47 | - | - | ||
| N(1)-C(3) | 1.47 | |||||
| En | N(2)-C(6) | 1.47 | - | - | ||
| N(1)-C(3) | 1.47 | |||||
| (CH3)2Sn(Et4en)Cl2 | N(12)-C(14) | 1.50 | Sn(1)-N(13) | 2.59 | - | |
| N(13)-C(17) | 1.50 | Sn(1)-N(1/2) | 2.63 | |||
| (CH3)2Sn(en)Cl2 | N(13)-C(17) | 1.49 | Sn(1)-N(13) | 2.40 | - | |
| N(12)-C(14) | 1.49 | Sn(1)-N(12) | 2.40 | |||
| (CH3)2Sn(Et4en) |
N(11)-C(15) | 1.52 | Sn(1)-N(11) | 2.33 | Sn(1)-O(48) | 2.46 |
| N(10)-C(12) | 1.52 | Sn(1)-N(10) | 2.33 | Sn(1)-O(46) | 2.43 | |
| (CH3)2Sn(en) |
N(11)-C(15) | 1.52 | Sn(1)-N(10) | 2.28 | Sn(1)-O(22) | 2.37 |
| N(10)-C(12) | 1.52 | Sn(1)-N(11) | 2.28 | Sn(1)-O(24) | 2.37 | |
| (CH3)2Sn(Et4en)(OH)2 | N(11)-C(15) | 1.50 | Sn(1)-N(10) | 2.76 | Sn(1)-O(48) | 1.99 |
| N(10)-C(12) | 1.50 | Sn(1)-N(11) | 2.83 | Sn(1)-O(46) | 1.99 | |
| (CH3)2Sn(en)(OH)2 | N(10)-C(12) | 1.49 | Sn(1)-N(10) | 2.48 | Sn(1)-O(24) | 1.99 |
| N(11)-C(15) | 1.49 | Sn(1)-N(11) | 2.48 | Sn(1)-O(22) | 1.99 | |
Table 6 Calculated N−C−C bond angles and the nitrogen atom Mulliken charges of the optimized structures of the ligands and complexes according to B3LYP/LanL2DZ level.
| Spices | N−C−C bond angle | Nitrogen atom charge | ||
|---|---|---|---|---|
| Et4en | C(6)-C(3)-N(1) | 115.5 | N(1) | -0.10 |
| C(3)-C(6)-N(2) | 115.9 | N(2) | -0.10 | |
| En | C(6)-C(3)-N(1) | 110.9 | N(1) | -0.63 |
| C(3)-C(6)-N(2) | 110.9 | N(2) | -0.63 | |
| (CH3)2Sn(Et4en)Cl2 | C(14)-C(17)-N(13) | 113.9 | N(12) | -0.26 |
| C(17)-C(14)-N(12) | 113.1 | N(13) | -0.26 | |
| (CH3)2Sn(Et4en) |
C(12)-C(15)-N(11) | 111.9 | N(10) | -0.39 |
| C(15)-C(12)-N(10) | 110.9 | N(11) | -0.39 | |
| (CH3)2Sn(en)Cl2 | C(17)-C(14)-N(12) | 109.5 | N(12) | -0.69 |
| C(14)-C(17)-N(13) | 109.5 | N(13) | -0.69 | |
| (CH3)2Sn(en) |
C(12)-C(15)-N(11) | 108.8 | N(10) | -0.76 |
| C(15)-C(12)-N(10) | 108.8 | N(11) | -0.76 | |
| (CH3)2Sn(Et4en)(OH)2 | C(12)-C(15)-N(11) | 114.7 | N(10) | -0.22 |
| C(15)-C(12)-N(10) | 113.5 | N(11) | -0.20 | |
| (CH3)2Sn(en)(OH)2 | C(15)-C(12)-N(10) | 110.2 | N(10) | -0.66 |
| C(12)-C(15)-N(11) | 110.2 | N(11) | -0.66 | |
Based on the DFT calculation, the polarity of the complexes, is much larger than that for the free ligands as indicated by the dipole moment of the complexes and the ligands (Table 6). On the other hand, the dipole moment of the hydroxo-complexes (CH3)2Sn(Et4en)(OH)2 and (CH3)2Sn(en)(OH)2 are lower than that of the corresponding dichloro-complexes (CH3)2Sn(Et4en)Cl2 (Table 7). This may be described on the assumption that the O-Sn(IV) bond is shorter than the Cl-Sn(IV) bond as given in Table 5.
Table 7 The calculated energies and dipole moments for of the ligands and complexes according to B3LYP/LanL2DZ level.
| Spices | Ea | HOMO | LUMO | ∆E | Dipole moment |
|---|---|---|---|---|---|
| Et4en | -504.9 | -4.84 | 2.76 | 7.60 | 0.69 |
| En | -190.5 | -5.51 | 2.70 | 8.21 | 1.05 |
| (CH3)2Sn(Et4en)Cl2 | -618.1 | -6.54 | -0.11 | 6.43 | 12.38 |
| (CH3)2Sn(en)Cl2 | -303.7 | -6.64 | 0.08 | 6.72 | 12.47 |
| (CH3)2Sn(Et4en)(OH)2 | -739.8 | -5.90 | 0.90 | 6.80 | 6.54 |
| (CH3)2Sn(en)(OH)2 | -425.4 | -5.88 | 1.39 | 7.27 | 7.33 |
a E: is the total energy (a.u.), HOMO: is the highest occupied molecular orbital (eV), LUMO: is the lowest unoccupied molecular orbital (eV), ∆E: ELUMO- EHOMO (eV) and dipole moment calculated (Debye).
The binding energy (BE) of each complex was calculated according to the following equation:
In equation 13, EAB, EA and EB is the energy of the complex, dimethyltin(IV) species and ligand respectively. The calculated BE (in kJ/mol) of the complexes (CH3)2Sn(Et4en)Cl2, (CH3)2Sn(en)Cl2, (CH3)2Sn(Et4en)(OH)2, (CH3)2Sn(en)(OH)2, (CH3)2Sn(Et4en)
Calculated infrared spectra of ligands and complexes
The experimental counterparts of the calculated IR spectra results are currently not available for comparison and it will be considered in our future work. Therefore, we will discuss the calculated IR spectra of the complexes from the theoretical point of view only. The calculated IR spectra of the free ligands and the complexes are shown in Fig. 7. There are no imaginary vibrational frequencies observed in all calculated spectra of the ligands and complexes. The assignments of the calculated vibrational frequencies of the free ligands and their complexes are listed in Table 8. The theoretical assignments are easily obtained by the visualization of the normal mode displacement vectors utilizing the GaussView program. The assignments listed in Table 8 are only for the vibrational modes of the amino group vibrations because they are the most sensitive to changes upon the metal-nitrogen interactions. Also, the Sn-N, Sn-OH, Sn-Cl and Sn-OH2 stretching vibrations are identified. In the spectra of the CH3)2Sn(en)Cl2, (CH3)2Sn(en)
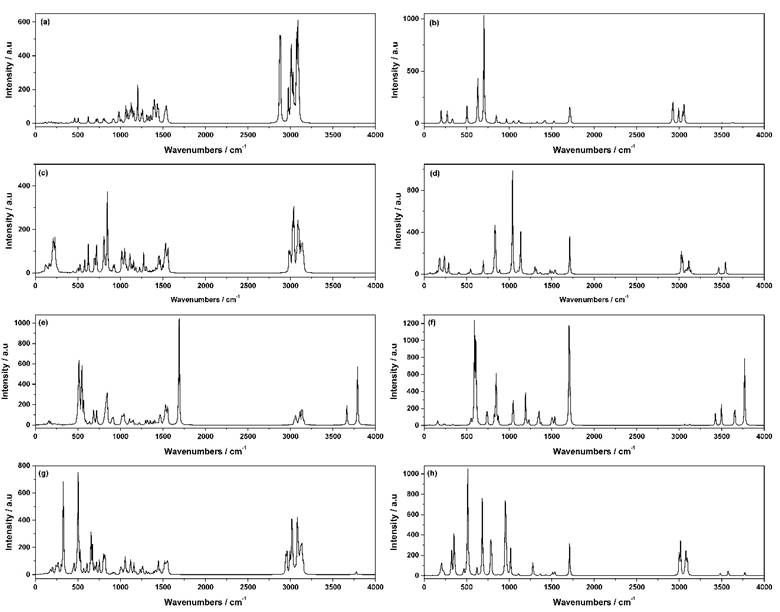
Fig. 7 Calculated Infrared spectra of the (a) Et4en ligand, (b) En ligand, (c) (CH3)2Sn(Et4en)Cl2 complex, (d) (CH3)2Sn(en)Cl2 complex, (e) (CH3)2Sn(Et4en)
Table 8 Calculated infrared absorption (cm−1) data for the ligands and complexes.
| Compounds | v(Sn‒Cl) | v (Sn‒OH) | v (Sn‒OH 2 ) | v (Sn‒N) | v as(Sn‒CH3) | v s(Sn‒CH3) | βs(NH2) | v (C‒N) | v as(C‒C‒N) |
|---|---|---|---|---|---|---|---|---|---|
| Et4en | - | - | - | - | - | - | - | 1021 | 1157 |
| En | - | - | - | - | - | - | 1670 | 1066 | 1172 |
| (CH3)2Sn(Et4en)Cl2 | 261 | - | - | 576612 | 537 | 471 | - | 1035 | 1133 |
| (CH3)2Sn(en)Cl2 | 260 | - | 285 | 552 | 490 | 1696 | 1027 | 1135 | |
| (CH3)2Sn(Et4en) |
- | - | 246242 | 510594 | 579 | 499 | - | 1036 | 1136 |
| (CH3)2Sn(en) |
- | - | 244 249 | 331 | 555 | 512 | 1696 | 1012 | 1064 |
| (CH3)2Sn(Et4en)(OH)2 | - | 519 524 | - | 568 603 | 541 | 466 | - | 1067 | 1131 |
| (CH3)2Sn(en)(OH)2 | - | 524 527 | - | 355 | 534 | 475 | 1691 | 1006 | 1131 |
a: Calculated value according to B3LYP/LANL2DZ level, ns: symmetric stretching, nas: asymmetric stretching n: stretching, βs: scissoring
Conclusion
The present investigation reports the complex formation equilibria of dimethyltin(IV) with N,N,N’,N’-tetraethylethylenediamine and its parent ethylenediamine. The results show the formation of 1:1 complex and the mono- and dihydroxo- species. The effect of solvent polarity on the stability of the complexes was discussed. The thermodynamic parameters of the complexes as ΔH° and ΔS° were determined and discussed. The displacement reaction of Et4en coordinated to dimethyltin (IV) by DNA constituents was investigated. IMP and GMP have the highest tendency to displace Et4en. The geometric parameters, bond angle, atomic charge and bond lengths were computed and interpreted. The results of the present study are expected to contribute to the chemistry of diorgantin (IV) as potential antitumours.











 nueva página del texto (beta)
nueva página del texto (beta)








