Introduction
Salmonella is globally recognized as the main food/waterborne etiological disease agent (WHO, 2018a). The Center for Disease Control and Prevention of the United States of America and the European Food Safety Authority report more than 1,000,000 and 100,000 non-typhoidal Salmonella infections annually, respectively (CDC, 2018a, EFSA, 2015), while in Mexico the number reaches approximately 70,000 cases of this condition each year (DGE, 2017).
Although this bacterium resides predominantly in the gastrointestinal tract of warm-blooded and coldblooded animals, its recognized ubiquity in natural environments, where it deploys survival strategies, allows it to prevail for prolonged periods in soils and sediments adapting itself to stressful conditions of temperature, pH, desiccation, osmotic and nutrimental stress, as well as predation facilitating its survival, environmental dispersion and the reach of new hosts (Donlan & Costerton, 2002; Tamagnini & Paraje, 2015).
Salmonella is persistent and shows a seasonal behavior, particularly when high temperatures and rainfalls are present, as those in tropical and subtropical regions where environmental conditions are ideal for the establishment of the bacteria (Simental & Martinez-Urtaza, 2008; Zhang et al., 2010; WHO, 2018b). Another important factor in the environmental persistence of Salmonella is its resistance to antibiotics, which increases the risk of infections in humans and animals due to the appearance of multi-resistant clones, further complicating the therapeutic treatment related to diseases and outbreaks (Angulo et al., 2004; Zaidi et al., 2012). In addition, the ability of Salmonella to form biofilms in different scenarios facilitates its adherence to survive on living or inert surfaces, a crucial step to infect a new host (Donlan & Costerton, 2002; Ledeboer & Jones, 2005; Steenackers et al., 2012).
The objective of this review was to describe the state that holds knowledge about the diversity of Salmonella and the factors that contribute to its persistence and prevalence in order to have a better understanding of its passage through the environment attempting to discern how this can impact this pathogen and therefore the epidemiology of the population of our country. This information will be useful to generate future research lines that will help in the design of strategies for the surveillance and control of this microorganism.
Result and Discussion
Epidemiology of Salmonella in Mexico
The genus Salmonella groups two species: bongori and enterica, the latter being divided into six subspecies: enterica, salamae, arizonae, diarizonae, houtenae and indica (Brenner et al., 2000). Of these, the subspecies enterica stands out, which is represented by more than 2600 serotypes that together generate millions of infections annually, ranging from acute gastroenteritis (nontyphoidal salmonellosis, NTS) to typhoid fever (Typhi and Paratyphi serotypes) in human beings (CDC, 2013). The main symptoms of NTS are inflammation of the gastrointestinal tract that is accompanied by nonbloody diarrhea, vomiting, nausea, headache, abdominal cramps and myalgia. In healthy individuals, the NTS is a self-limiting infection, with a recovery period from 2 to 4 days; however, susceptible groups of the population, such as children, elderly and immunocompromised, are vulnerable to suffer from serious conditions that can even cause death (CDC, 2018b). Of the 2,600 serotypes Typhimurium and Enteritidis lead the reports of diarrheal outbreaks, for which they are considered the main responsible for the NTS (CDC, 2018b).
The epidemiological surveillance systems are an effective tool to know with relative certainty the incidence of diarrheal outbreaks generated by the consumption of contaminated food, as well as the impact on the morbidity and mortality of the population. Unfortunately, these systems only work properly in developed countries, while in developing countries their functionality is deficient. This dysfunction is associated with the population’s lack of interest in seeking medical attention in case of a diarrheal event, probably due to the general understanding of the self-limiting behavior commonly exhibited by these infections, and the economic burden involved in the clinical and treatment processes, as well as the deficiencies in public health systems that demotivate the patient to demand their rights as a beneficiary of the system.
Health authorities in Mexico have made a great effort to strengthen the national epidemiological surveillance system (Sistema Nacional de Vigilancia Epidemiológica, SINAVE), which is responsible for collecting information on epidemiological events, including cases of salmonellosis, which are reported weekly, complying with the Standard Mexican Official NOM-017-SSA2-1994.
In this sense, the SINAVE shows in the period from 1984 to 2017 a considerable increase of typhoid fever going from 7,629 to 45,280 cases; and for paratyphoid fever/other salmonellosis from 31,943 to 104,471 cases, respectively (Figure 1 and 2). These statistical data show Sinaloa and Tamaulipas as the states with the highest rate of typhoid fever with 12.9 % and 10.25 %, respectively. In the case of paratyphoid fever and other salmonellosis, Chiapas (12.99 %), Veracruz (9.37 %) and Tabasco (8.31 %) are the states with the highest number of cases (INEGI, 2017) registered in the months of March to October (DGE, 2017). This information exhibits an increase of approximately double the cases of salmonellosis in the female population in relation to the male gender (DGE, 2017). These findings agree with the dynamics worldwide and specifically with that cited by Reller et al., (2008) where an increase of salmonellosis in women in the years comprised from 1968 to 2000 is reported. The possible causes of this global and national trend are associated with the following factors: the female Mexican population is greater than the male population, biological susceptibility, and higher frequency of exposure with potential sources of contamination.
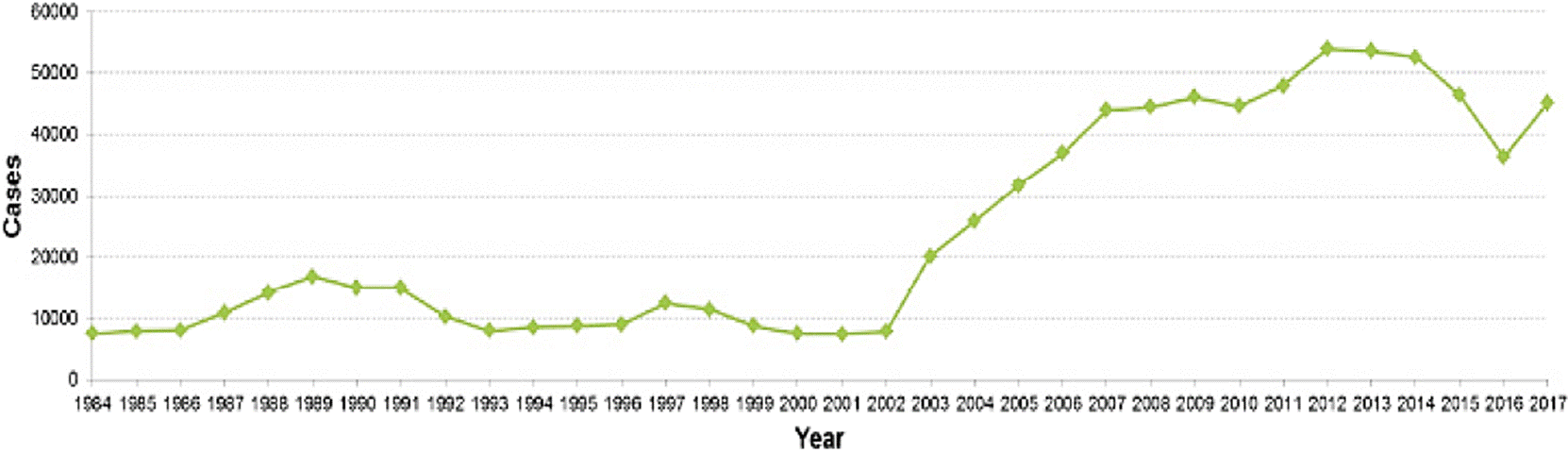
Figure 1 Prevalence of Typhoid Fever in Mexico during the period from 1984 to 2017 (Anuario de Morbilidad 1984-2017. DGE, 2017).
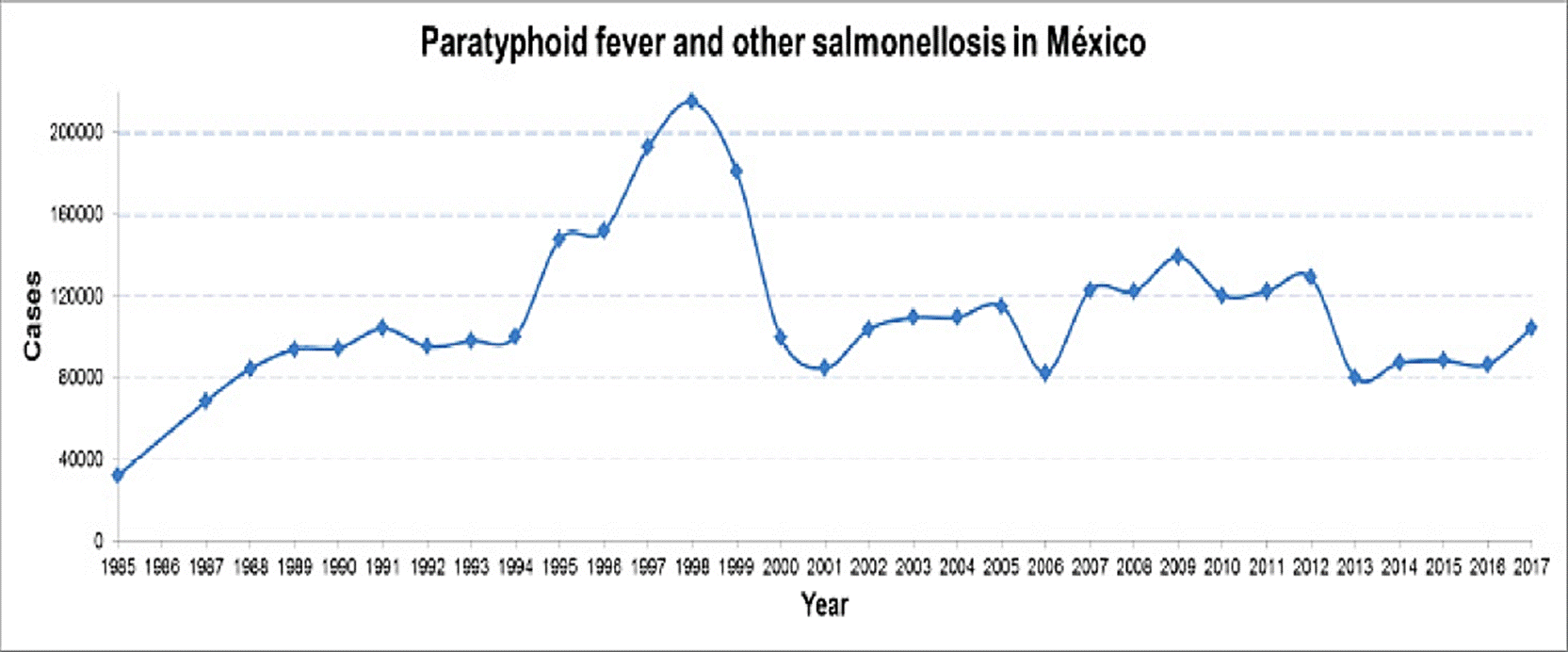
Figure 2 Prevalence of Paratyphoid fever and other salmonellosis in Mexico during the period from 1984 to 2017. (Anuario de Morbilidad 1984 -2017. DGE, 2017).
In this review from 1968 to 2018, a wide range of Salmonella serotypes isolated from the environment, food, humans and animals were detected in Mexico, highlighting Enteritidis, Typhimurium, Anatum, Agona, Meleagridis, Oranienburg, Derby, Infantis, Ohio and Havana as the most predominant serotypes (Table 1); This predominance serotype pattern agrees to a large extent with the study carried out by Gutiérrez-Cogco et al., (2000) where it presents Typhimurium, Enteritidis, Derby, Agona and Anatum as the main Salmonella serotypes circulating in the country in the period from 1972 to 1999, isolated from diverse sources (human, environment, food).
Table 1 Diversity of Salmonella serotypes in Mexico isolated from different sources from 1968 to 2018.
| Isolation origin | ||||||||
|---|---|---|---|---|---|---|---|---|
| Serotype | Animal | Human | Enviroment | Food of animal origin | Food of plant origin | Total | % | References* |
| Enteritidis | 7 | 190 | 1 | 53 | 6 | 257 | 6.61 | 27, 2, 46, 44, 11, 10, 35, 19, 42, 36, 22, 15 |
| Typhimurium | 17 | 63 | 38 | 100 | 29 | 247 | 6.52 | 27, 2, 46, 34, 44 |
| Anatum | 25 | 38 | 18 | 120 | 4 | 205 | 5.41 | 27, 24, 18, 2, 46, 34, 44, 1, 29, 41,
10, 35, 13, 40, 36 |
| Agona | 3 | 67 | 5 | 106 | 8 | 189 | 4.99 | 27, 46, 44, 27, 14,
29, 10, 35, 17, 40, 36, 15 |
| Meleagridis | 1 | 41 | 0 | 118 | 0 | 160 | 4.22 | 27, 46, 44 |
| Oranienburg | 32 | 43 | 50 | 2 | 0 | 127 | 3.35 | 38, 27, 2, 43, 46, 39, 19, 29, 14, 17 |
| Derby | 14 | 4 | 0 | 66 | 0 | 84 | 2.21 | 38, 24, 2, 44, 41, 13, 17, 32 |
| Infantis | 4 | 35 | 7 | 36 | 0 | 82 | 2.16 | 24, 18, 2, 34, 44, 28, 29, 41, 46, 13, 40 |
| Ohio | 0 | 67 | 0 | 2 | 0 | 69 | 1.82 | 34, 13 |
| Havana | 2 | 8 | 0 | 49 | 0 | 59 | 1.55 | 24, 44, 13 |
| Reading | 0 | 13 | 0 | 38 | 0 | 51 | 1.34 | 44, 46, 40 |
| Albany | 1 | 15 | 0 | 29 | 0 | 45 | 1.18 | 27, 44, 46, |
| Saintpaul | 11 | 0 | 27 | 6 | 0 | 44 | 1.16 | 27, 2, 28,14, 13, 17,18 |
| Arizonae | 24 | 0 | 0 | 1 | 12 | 37 | 0.97 | 30, 35, 36 |
| Adelaide | 0 | 13 | 0 | 23 | 0 | 36 | 0.95 | 44, 41, 40 |
| Give | 16 | 0 | 17 | 2 | 0 | 35 | 0.92 | 27, 2, 28, 14, 1, 29 |
| Wourthington | 0 | 0 | 0 | 30 | 0 | 30 | 0.79 | 46, 13 |
| Seftenberg | 15 | 0 | 1 | 11 | 0 | 27 | 0.71 | 2, 28, 13, 17, 32, 40 |
| Newport | 3 | 11 | 4 | 4 | 0 | 22 | 0.58 | 27, 2, 46, 34, 28, 44 |
| Minnesota | 9 | 0 | 8 | 3 | 0 | 20 | 0.52 | 27, 2, 28, 29, 13 |
| Typhi | 0 | 0 | 6 | 11 | 3 | 20 | 0.52 | 4, 32, 36, 21 |
| Muenchen | 4 | 9 | 0 | 5 | 0 | 18 | 0.47 | 27, 2, 46, 44 |
| Weltevreden | 9 | 0 | 7 | 0 | 1 | 17 | 0.44 | 39, 27, 28,14 |
| Choleraesuis | 3 | 0 | 0 | 2 | 12 | 17 | 0.44 | 2, 35, 40, 36 |
| London | 1 | 0 | 0 | 15 | 0 | 16 | 0.42 | 2, 32 |
| Hadar | 12 | 0 | 0 | 0 | 0 | 12 | 0.31 | 2 |
| Sandiego | 9 | 0 | 3 | 0 | 0 | 12 | 0.31 | 38, 24, 28,14 |
| Braenderup | 5 | 0 | 1 | 6 | 0 | 12 | 0.31 | 38, 28, 47 |
| Poona | 5 | 0 | 7 | 0 | 0 | 12 | 0.31 | 18, 2, 28, 18 |
| Stanleyville | 0 | 3 | 1 | 7 | 0 | 11 | 0.29 | 44, 39, 46 |
| Muenster | 1 | 0 | 5 | 4 | 0 | 10 | 0.26 | 24, 28, 41 |
| Brandenburg | 3 | 0 | 0 | 6 | 0 | 9 | 0.23 | 2, 46 |
| Gallinarum | 0 | 0 | 0 | 1 | 8 | 9 | 0.23 | 35, 36 |
| Bredeney | 1 | 0 | 0 | 7 | 0 | 8 | 0.21 | 24, 17, 40 |
| Heidelberg | 7 | 0 | 0 | 0 | 0 | 7 | 0.18 | 2 |
| Duesseldorf | 6 | 0 | 0 | 0 | 0 | 6 | 0.16 | 2 |
| Bovismorficans | 2 | 0 | 2 | 2 | 0 | 6 | 0.16 | 2, 28, 17 |
| Kentucky | 4 | 0 | 0 | 2 | 0 | 6 | 0.16 | 24, 2,10 |
| Azteca | 0 | 0 | 0 | 6 | 0 | 6 | 0.16 | 41 |
| Cerro | 0 | 0 | 0 | 6 | 0 | 6 | 0.16 | 46 |
| Lomita | 0 | 0 | 0 | 6 | 0 | 6 | 0.16 | 32 |
| Cannstatt | 0 | 0 | 0 | 6 | 0 | 6 | 0.16 | 46, 32 |
| Panama | 0 | 6 | 0 | 0 | 0 | 6 | 0.16 | 44 |
| Montevideo | 2 | 0 | 4 | 0 | 0 | 6 | 0.16 | 27, 2, 28 |
| Luciana | 3 | 0 | 2 | 0 | 0 | 5 | 0.13 | 27, 28 |
| Pomona | 0 | 0 | 5 | 0 | 0 | 5 | 0.13 | 28 |
| Houtenae | 0 | 0 | 0 | 1 | 4 | 5 | 0.13 | 35 |
| Vejle | 0 | 0 | 4 | 0 | 0 | 4 | 0.10 | 39 |
| Javiana | 1 | 0 | 2 | 1 | 0 | 4 | 0.10 | 27, 28, 32 |
| Edimburg | 0 | 0 | 0 | 1 | 3 | 4 | 0.10 | 35, 36 |
| Tennessee | 2 | 0 | 0 | 1 | 0 | 3 | 0.08 | 2, 32 |
| Soahanina | 2 | 0 | 1 | 0 | 0 | 3 | 0.08 | 27, 28 |
| Schwarzengrund | 0 | 0 | 0 | 3 | 0 | 3 | 0.08 | 13 |
| Acquatoria | 0 | 0 | 0 | 3 | 0 | 3 | 0.08 | 13 |
| Suberu | 0 | 0 | 3 | 0 | 0 | 3 | 0.08 | 39 |
| Urbana | 0 | 0 | 3 | 0 | 0 | 3 | 0.08 | 39 |
| Paratyphi | 0 | 0 | 3 | 0 | 0 | 3 | 0.08 | 4 |
| Degania | 2 | 0 | 0 | 0 | 0 | 2 | 0.05 | 30 |
| Binza | 0 | 0 | 0 | 2 | 0 | 2 | 0.05 | 13 |
| Tonev | 0 | 0 | 1 | 1 | 0 | 2 | 0.05 | 39 |
| Salamae | 0 | 0 | 0 | 0 | 2 | 2 | 0.05 | 36 |
| Sundsvall | 0 | 0 | 2 | 0 | 0 | 2 | 0.05 | 28 |
| Galiema | 0 | 0 | 2 | 0 | 0 | 2 | 0.05 | 39 |
| Othmarshem | 0 | 0 | 2 | 0 | 0 | 2 | 0.05 | 39 |
| Soerenga | 0 | 0 | 2 | 0 | 0 | 2 | 0.05 | 39 |
| Gaminara | 1 | 0 | 0 | 0 | 0 | 1 | 0.03 | 27 |
| Cayar | 1 | 0 | 0 | 0 | 0 | 1 | 0.03 | 27 |
| Rubislaw | 1 | 0 | 0 | 0 | 0 | 1 | 0.03 | 24 |
| Shamba | 1 | 0 | 0 | 0 | 0 | 1 | 0.03 | 30 |
| Selby | 1 | 0 | 0 | 0 | 0 | 1 | 0.03 | 30 |
| Bere | 1 | 0 | 0 | 0 | 0 | 1 | 0.03 | 30 |
| Bunnik | 1 | 0 | 0 | 0 | 0 | 1 | 0.03 | 30 |
| Drypool | 0 | 0 | 0 | 1 | 0 | 1 | 0.03 | 17 |
| Newlands | 0 | 0 | 0 | 1 | 0 | 1 | 0.03 | 17 |
| Winnipeg | 0 | 0 | 0 | 1 | 0 | 1 | 0.03 | 39 |
| Pullorum | 0 | 0 | 0 | 0 | 1 | 1 | 0.03 | 36 |
| Bongor | 0 | 0 | 0 | 0 | 1 | 1 | 0.03 | 36 |
| Kiambu | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 28 |
| Texas | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 28 |
| Amherstiana | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 39 |
| Augusten- burg | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 39 |
| Breda | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 39 |
| Bulovka | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 39 |
| Coeln | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 39 |
| Corvallis | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 39 |
| Djugu | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 39 |
| Nchanga | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 39 |
| Nitra | 0 | 0 | 1 | 0 | 0 | 1 | 0.03 | 39 |
| Stanley | 0 | 0 | 1 | 0 | 0 | 1 | 0.30 | 93 |
| Others | 612 | 187 | 43 | 711 | 76 | 1,629 | 41.27 | 12, 27, 24, 30, 38, 2, 46, 28, 18, 14, 39, 4, 24 , 37, 20, 35, 5, 39, 8, 10, 32, 42, 40, 20, 31, 36, 1, 22, 15, 9, 16, 3, 47, 25 |
*1.Acedo-Félix et al., 2009; 2. Alaniz-de la O et al., 1999; 3. Alcázar-Montañez et al., 2006; Becerra-Tapia & Botello, 1995; 5. Bello-Pérez et al., 1990; 6. Benavides-Plasencia et al., 2005; 7. Bessudo et al., 1973; 8. Camacho et al., 2010; 9. Cerna-Cortes et al., 2013; 10. Charles-Hernández et al., 2007; 11. Chávez-de la Peña et al., 2001; 12. Cueto-Medina et al., 2015; 13. Escartin et al., 1999; 14. Estrada-Acosta et al., 2014; 15. Estrada-García et al., 2004; 16. Félix-Fuentes et al., 2005; 17. Fernández-Escartín & Torres, 1996; 18. Figueroa et al., 2005; 19. Gallegos-Robles et al., 2008; 20. Gallegos-Robles et al., 2009; 21. García-Gómez et al., 2002; 22. Gómez-Aldapa et al., 2014; 23. González-Cortés et al., 1974; 24. Grabert, 1968; 25. Hernández et al., 2007; 26. Hernández-Domínguez et al., 2008; 27. Jiménez et al., 2011; 28. Jiménez et al., 2014; 29. López-Cuevas et al., 2009; 30. Martínez et al., 1999; 31. Morales-Hernández et al., 2009; 32. Nayarit-Ballesteros et al., 2016; 33. Olarte & Galindo, 1973; 34. Paniagua-Contreras et al., 2008; 35. Quiñonez-Ramírez et al., 2000; 36. Quiroz-Santiago et al., 2009; 37. Rubio et al., 2013; 38. Silva-Hidalgo et al., 2012; 39. Simental & Martínez-Urtaza, 2008; 40. Talavera et al., 2011; 41. Torres et al., 2011; 42. Varela-Guerrero et al., 2013; 43. Vázquez-Garcidueñas et al., 2014; 44. Zaidi et al., 2006; 45. Zaidi et al., 2007; 46. Zaidi et al., 2008; 47. Zaidi et al., 2012.
This type of information exposes an important area of opportunity to strengthen the epidemiological systems in the study of Salmonella in Mexico, since one of the main limitations at the moment of conducting epidemiological surveillance lies in the techniques used for the necessary monitoring for this process; the methodologies used are usually laborious, expensive, nonspecific and timeconsuming to obtain results, and finally the search is restricted to only two serotypes (Typhi and Paratyphi). For this reason, in recent years, the adoption of modern strategies that address the limitations presented by conventional methods and offer quick, reliable, simple or automated and economic alternatives has been promoted among the scientific community. The whole genome sequencing is a tool that is revolutionizing the microbiological analysis at present, allowing to obtain relevant information of specific individuals in record time by means of detection, monitoring, characterization and analysis of pathogenic microorganisms through comparative and functional genomics, also applied in the reconstruction of the dispersion dynamics of the mentioned microorganisms and their evolutionary history.
The innovative genomic tools will revolutionize epidemiological surveillance, offering useful tools to monitor, prevent and strategy design for the control of pathogenic microorganisms responsible for epidemiological outbreaks. This in turn will have a strong impact on the safety along the food chain production and public health.
Predisposing factors for the prevalence of Salmonella
Extreme environmental conditions such as heat waves, high rainfall and intrinsic characteristics of the pathogen that include the biofilm formation and antimicrobial resistance in Salmonella clones can deal in favorable conditions for the establishment, persistence and dispersion of bacterium in ecological niches (Angulo et al., 2004; Ledeboer & Jones, 2005; Akil et al., 2014).
Climate change
Climate change is defined as the variation of the earth’s weather over an extended period of time, which is related to natural events or direct/indirect human activities, affecting the parameters of temperature, rainfall and cloudiness (WHO, 2018b). The atmosphere is composed of various gases, which contribute to maintain solar heat around the earth, dealing in the suitability of the planet. Among these gases are carbon dioxide, nitrous oxide and methane, which are called greenhouse gases and their levels have been increasing due to different anthropogenic activities. One of these gases, CO2, is the one that most traps heat in the lower layers of the atmosphere, this has generated an increase of 0.85 °C in the average temperature of the planet in the last century, allowing global warming to occur and concomitant to climate change (WHO, 2018b).
Human health can be severely affected by climate change by its influence on the interaction between human and pathogens, since different climatic phenomena can promote the occurrence of airborne, foodborne and waterborne infections associated with pathogenic microorganisms, especially between regions and susceptible populations (Wu et al., 2016). This has been evidenced by Akil et al., (2014) and Jiang et al., (2015) who report that concomitantly high temperatures and rainfall intensity are factors that have a direct relationship with the increase of outbreaks associated with pathogens such as Salmonella.
This kind of climatic variations has showed a direct impact in some regions of the Mexican territory, since climatic events as hurricanes are more frequent and intense, and generate serious floods, especially in zones located near the Atlantic Ocean and the Northeastern coastal areas (CONAGUA, 2013). An example of this are the high incidence of typhoid fever in states such as Sinaloa and Tamaulipas (DGE, 2017) and paratyphoid/ other salmonellosis in Chiapas, Veracruz and Tabasco; these states have similar temperature and rainfall and are hit by cyclones and hurricanes (CONAGUA 2013; INEGI, 2017), being the months of march to October where the greatest number of cases of salmonellosis (DGE, 2017).
These data concur with the geographical distribution of typhoid fever, placing it in developing countries, which converge geo-spatially in the tropic of cancer, having a tropical climate (Lee et al., 2016). However, although Mexico is located within this tropical climate, its territory is divided by microclimates, which are distinguished by unique environmental characteristics offering specific niches, which can signify a challenge to overcome during the environmental phase of any microorganism.
The diversity of microclimates and their effect on the presence of pathogens such as Salmonella are exposed in contrasting studies such as those by Simental & Martínez-Urtaza (2008) and Jiménez et al., (2014) who showed the influence of the geographical and seasonal distribution; the first associating Salmonella with periods of rain in places such as Bahía de Todos Santos, Baja California, Mexico; while the second, detecting the bacteria consistently regardless of the season of the year in rivers of Sinaloa. The divergence between these two studies may be due to differences in microclimates in these regions, due to the ecoregions in which the two states are located, identifying the California-Mediterranean for the case of Baja California and the warm-dry forests for Sinaloa (INEGI, 2017).
To face climate change it is necessary the effort and joint coordination of the government, civil society and the health sector to formulate strategies and sensitize the population to minimize global warming and its secondary effects, as well as to establish timely strategies for the prevention of infectious diseases. This mainly as a result of the increasing world population, since only Mexico has a little more than 112 million inhabitants (INEGI, 2017), leading to the intensification of the adverse effects of climate change, since anthropogenic activities are a main cause of this phenomenon.
Biofilm formation
Once released in the environment, Salmonella is exposed to non-host conditions, which can induce the bacterium to activate adaptation mechanisms (Winfield & Groisman, 2003, Martínez-Urtaza et al., 2004). One the most common mechanisms of adaptation used by bacteria in the environment, is the formation of biofilms, which are bacterial associations surrounded by polymeric matrix adhered to living or non-living surfaces (Costerton et al., 1999), and can be formed in three interfaces: liquid-air, solid-air and solid-liquid (Constantin, 2009; Steenackers et al., 2012). These structures confer protection against stressful or hostile environmental factors, besides representing a source of constant and persistent bacterial dissemination (Donlan & Costerton, 2002, Jensen et al., 2010). Salmonella biofilms are constituted by water (70-97 %), nucleic acids, exopolysaccharides such as cellulose and colanic acid, as well as proteins fimbria curli and BapA (Steenackers et al., 2012). Several studies have shown that bacteria, depending on environmental conditions and the surface where they adhere, produce different exopolysaccharides and proteins as components of the biofilm matrix, resulting in a different composition those biofilms formed in animal tissues, plants or inert surfaces (glass, plastic, stainless steel) (Costerton et al., 1999; Boddicker et al., 2002; Steenackers et al., 2012). Undoubtedly, the formation of biofilms in any space (environment, host) where it is formed, improves the ability of Salmonella to survive (Ledeboer & Jones, 2005).
In this sense, in Mexico Medrano-Félix et al. (2018a) demonstrated that the capacity of environmental Salmonella serotypes prevalent in an agricultural region to develop biofilms in the liquid-air interface depends on the levels of pH and salinity of water. Silva-Hidalgo et al., (2016) detected the bapA gene in serovars of Salmonella enterica subsp. enterica isolated of animals in captivity, highlighting the importance of this gene in the conformation of biofilms and how it favors the resistance of this bacterium in natural habitats. Other studies demonstrated the biofilm forming ability of Salmonella Poona and Michigan in bark cantaloupe (Annous et al., 2005) and Salmonella Typhimurium and Javiana in cactus (Opuntia ficus-indica), water and soil (De los Santos et al., 2012) showing an increase in the resistance of this pathogen to disinfectants. The joint analysis of these works exhibits the life cycle of Salmonella wrapped by the protection of these survival structures that allow it to remain viable when exposed to different scenarios. Although these are efforts to explain the role of exopolymeric matrices in the biology of Salmonella strains circulating in the country, there is still a need to widen the studies in this topic for a better understanding on how predominant environmental factors in Mexico regulate and influence the formation/composition of biofilms, and whether these interaction potentiate the success to reaching a new host.
Antimicrobial resistance (AMR)
AMR is the ability of microorganisms to prevent antibiotics from acting against them (WHO, 2016). One of the main causes that detonate resistance to antimicrobials is their indiscriminate use (Angulo et al., 2004). The success of AMR bacteria is due to the ability to mutate and exchange genetic material between different bacterial species.
A significant number of research groups have focused their efforts on characterizing the different levels of resistance that may occur in different Salmonella serotypes ranging from uni-resistance to multiple resistance to antibiotics such as ampicillin, chloramphenicol, streptomycin, sulfonamides and tetracycline (Bessudo et al., 1973; Olarte & Galindo, 1973; González-Cortes et al., 1974; Alaniz-de la O et al., 1996; Miranda et al., 2009; Camacho et al., 2010; Talavera et al., 2011; Jiménez et al., 2011; Pérez-Montaño et al., 2012; Nayarit-Ballesteros et al., 2016; Aguilar-Montes de Oca et al., 2017), all of these drugs are used as first-line therapy in the treatment of conditions caused by Salmonella.
This was exposed in a case study of a gastrointestinal outbreak due to Salmonella Oranienburg, which occurred in a Mexican prison, where a high diversity of resistance patterns was observed (17 AMR profiles), among which resistance to ampicillin, carbenicillin and chephalothin was predominantly found (Vázquez-Garcidueñas et al., 2014). AMR is not limited to pathogenic serotypes typical of humans, which are commonly exposed to antimicrobials, but is also observed in strains that predominate in nonhost environments, as described by López-Cuevas et al., (2009) who detected a high resistance to tetracycline in Salmonella Typhimurium (12/13) isolated from water in the valley of Culiacan, Sinaloa. Subsequently, Lugo-Melchor et al., (2010), after more detailed studies of these strains by means of PCR and PFGE, identified the tet(A) gene (which confers tetracycline resistance) in the Tn1721 transposon in the genomic DNA. The development of AMR in Salmonella during its passage through the environment can be mediated by genetic changes promoted by the exposure of bacterial populations to drugs used in an intensive and indiscriminate manner in both agricultural and livestock activities.
Due to the challenge that bacterial control through the use of drugs implies today, it is necessary to find alternatives that are available to the entire population and that guarantee lasting protection to reduce the use of antibiotics and their adverse effects, generating alternative lines of research, such as bacteriophages (widely used as biological control and phage therapy) or the development of vaccines for other serotypes other than Salmonella Typhi.
Diversity of Salmonella in non-host environments
The occurrence of non-typhoidal salmonellosis cases in humans is traditionally related to the consumption of animal origin products contaminated with Salmonella (WHO, 2018a); however, the number of outbreaks associated with the consumption of fresh produce has increased in recent years (CDC, 2018b). Animal excretion constitutes the main route for enteric pathogens release from the host to the environment. These wastes may contaminate rivers, lakes and other wetlands when deposited directly into water bodies; other deposition vias are rainfall runoff and anthropogenic activities such as agriculture and cattle raising; or their release through drainage outlets from cities and other human settlements. This is the reason why water is considered an important vehicle for the dispersion of Salmonella in the environment (Winfield & Groisman, 2003).
Knowing the mechanisms and understanding the dispersion dynamics of Salmonella in the environment requires meticulous studies that reconstruct the historical spreading of the microorganism through time and space, considering both geographic areas and sources of isolation, along with the diversity profiles of Salmonella populations and establishing the specific role played by each of the factors during Salmonella passage from the environment to the next host. As the knowledge about these processes and biological interactions is deepened, the design and implementation of efficient strategies for the prevention, diagnosis and control of Salmonella will be improved, with the aim of avoiding epidemiological outbreaks and minimizing the impact of Salmonella on public health.
Next, we describe the Salmonella diversity found by different authors in a variety of matrices such as water, surfaces and food, as well as their possible implications in the dissemination of the bacterium.
Water, soil and inert surfaces
Non-treated irrigation water and soils enriched with compost are considered the main sources of contamination that may cause the loss of food safety. Water is a vehicle that can magnify point contamination and generate Salmonella dispersion in a large extent (Martínez-Urtaza et al., 2004, Levantesi et al., 2012).
The presence of Salmonella in irrigation water is part of deficient agricultural and manufacturing practices that can increase the likelihood of contaminating food in agricultural packinghouses or even the production in the field. Studies on the safety of melon, pepper, tomato and cilantro in the Mexican states of Coahuila, Michoacán, Guerrero, Sinaloa and Sonora agree that irrigation water is a vehicle for Salmonella contamination (Gallegos-Robles et al., 2008; Figueroa et al., 2005; Hernández-Domínguez et al., 2008; López-Cuevas et al., 2009; Acedo-Félix et al., 2009; Morales-Hernández et al., 2009; Estrada-Acosta et al., 2014). The diversity of serotypes found in these studies can be attributed to the fact that water bodies are outdoors, so the incorporation of Salmonella species is multifactorial including wastewater (industrial or domestic), wild animals feces (mammals, reptiles and birds) as well as floods and runoff promoting eutrophication processes, which is beneficial for the microorganism (Simental and Martínez-Urtaza, 2008; Jiménez et al., 2014).
In Mexico, the most common Salmonella serotypes isolated from water, crop soil and packaging surfaces are Oranienburg, Typhimurium, Saintpaul, Anatum and Give, of which the main sources of isolation were river water, irrigation channels and streams. The isolation of these serotypes involved the use of several techniques, such as Polymerase Chain Reaction (PCR), serotyping (Figueroa et al., 2005; Simental & Martínez-Urtaza, 2008; Hernández-Domínguez et al., 2008; Morales-Hernández et al., 2009; López-Cuevas et al., 2009), Restriction Fragment Length Polymorphism (RFLP) (Gallegos-Robles et al., 2008), and Pulsed-Field Gel Electrophoresis (PFGE), aiming the diversity establishment based on genetic relationships between isolates from different sources, incorporating georeferencing in order to obtain differential geographic patterns (Estrada-Acosta et al., 2014; Jiménez et al., 2014). In these last two studies, the existing intraserotype diversity in an ecological niche was evidenced, reaching up to 22 different profiles from a population of 58 strains comprised of seven serotypes; while some profiles showed to be exclusive of a particular point, some others were distributed in different points of that space.
Although, the PFGE technique was a standard widely used by the CDC and contributed to the knowledge establishing the microbial genetic diversity based on its scope, nowadays according to the current requirements, this tool does not suffice in order to discriminate and establish a reliable temporal-space relationship among strains, because of limited capacity to distinguish between two closely genetically related individuals. This is the reason is why we suggest the use of more modern and efficient strategies such as Whole Genome Sequencing.
Metabolic flexibility is among the survival abilities that Salmonella employs when in the environment, since it has been shown that once this pathogen incorporates into water bodies it is not limited by the scarcity and type of available carbon sources. Medrano-Félix et al., (2018b) reported the ease of adaptation of clinical origin Salmonella serotypes by consuming a wide variety of carbon sources. Similarly, environmental serotypes showed greater metabolic activity, but the carbon sources diversity used was narrow; besides a higher effect in adhesion, invasion and cell viability was observed, when tested in epithelial cells demonstrating a greater pathogenic capacity (Estrada-Acosta et al., 2017). These findings suggest the study and surveillance of Salmonella should not be limited to human-specific serotypes.
On the other hand, studies of comparative genomics between Salmonella isolates obtained from surface water samples and feces from asymptomatic domestic animals showed that the genetic diversity among the different isolates is minimal, suggesting a close relationship between local isolates from both sources, and providing evidence that supports the theory that animals are an important source for Salmonella contamination to water bodies and that they, in turn, help in the transmission of the bacteria to new hosts (Jiménez et al., 2011; 2014).
Food of plant origin
Irrigation water contaminated with Salmonella has been associated with epidemiological outbreaks; this may be caused by inefficient disinfection processes or inadequate hygienic practices (in the field and packaging) resulting in the presence of this microorganism in fresh consumption food (Levantesi et al., 2012). In Mexico, several studies show the presence of Salmonella in fresh and processed vegetables, such as lettuce and coriander (García-Gómez et al., 2002), cantaloupe melon and chili (Morales-Hernández et al., 2009; Gallegos-Robles et al., 2008, 2009), tomato (Estrada-Acosta et al., 2014), bean sprouts (Cerna-Cortes et al., 2013), parsley, broccoli, cauliflower, spinach (Quiroz-Santiago et al., 2009), fruit salad (Félix-Fuentes et al., 2005), sauces for tacos (Estrada-García et al., 2004), corn tortillas (Gómez-Aldapa et al., 2013), fresh beet juice (Gómez-Aldapa et al., 2014) among others. Melon, parsley and coriander showed the highest Salmonella counts, while Typhimurium, Arizonae, Choleraesuius and Enteritidis were the most prevalent serotypes.
Data shown in these studies are consistent with investigations of outbreaks occurred in the United States from which an 11 % were caused by Salmonella serotypes Enteritidis and Typhimurium, being fresh leafy vegetables the food most commonly associated to (Painter et al., 2013; Herman et al., 2015); in addition to these reports, it is necessary to point out that Mexico has been involved in Salmonella outbreaks in fresh products such as papaya, cucumber, melon, mango, tomato (CDC, 2018b), therefore, it is necessary to focus more on the study of this pathogen in fresh produce, since the data reported in Mexico are scarce.
Food of animal origin
Domestic animals are the largest reservoirs of Salmonella, whose production of meats and derivatives are the most reported vehicles in outbreaks of salmonellosis transmitted by food (WHO, 2018a). Since the 90s, Salmonella was isolated in Mexico in 42.76 % of foods of animal origin, mainly of pork, beef and chicken (Charles-Hernández et al., 2007; Zaidi et al., 2006; Hernández et al., 2007; Zaidi et al., 2008; Camacho et al., 2010; Rubio et al., 2013; Varela-Guerrero et al., 2013; Nayarit-Ballesteros et al., 2016), followed by chorizo, longaniza and jerky (Bello-Pérez et al., 1990; Escartin, 1999; Charles-Hernández et al., 2007; Torres et al., 2011). In 2005, Mancera et al., 2005, isolated a low percentage (0.25 %) of Salmonella Enteritidis from the egg yolk. This is relevant because the main food product of the Mexican basic basket is the egg and its contamination of origin could mean an important public health risk. Another important source of Salmonella is marine products, which are commonly consumed raw. The diversity of Salmonella serotypes in fish ceviche (Fernández-Escartín & Torres-Vitela, 1996), clams (Quiñonez-Ramírez et al., 2000) and molluscs (Simental & Martínez-Urtaza, 2008) sold in fixed and mobile establishments shows a higher incidence of this pathogen during the warmer seasons.
For all the above, the contamination of food by Salmonella is of paramount importance to the Ministry of Health in Mexico, so there are standards for food control that specify the minimum requirements of good hygiene practices that should be observed in the process of food and its raw materials in order to avoid its contamination. Even with these measures there are a high number of Salmonella serotypes in foods of animal origin, evidencing the high risk of consumption of these products under a deficient cooking process.
Diversity of Salmonella in host enviroments
In general Salmonella enterica can be found in a variety of vertebrates, including humans, pets, wild animals, calves, poultry, pigs, sheep, wild birds and reptiles (CDC, 2018b). This bacterium is transported asymptomatically in the intestine or gallbladder of many animals and excreted continuously or intermittently through the stool (Gunn et al., 2014).
Animals
For more than half a century, Salmonella has been reported in animals in Mexico. In this review, animals occupy the second place as a source of isolation (23.42 %) of the bacteria (Table 1), of which cattle (10.14 %), poultry (5.63 %) and swine (3.38 %) had the highest prevalence, followed by sheep, dogs, American cockroaches and iguanas with 2.02, 0.56, 0.33 and 0.11 %, respectively. The main serotypes found were Oranienburg, Anatum, Arizonae and Typhimurium (Grabert, 1968; Figueroa et al., 2005; Jiménez et al., 2011; Cueto-Medina et al., 2015).
On the other hand, studies with animals in captivity have shown the presence of Salmonella; Martínez et al., (1999) studied feces of different species of reptiles in a serpentarium of the city of Puebla, finding 7.77 % of Salmonella isolates. Likewise, Silva-Hidalgo et al., (2012) reported the presence of this bacterium in 11.6 % of the stool samples of captive mammals, birds and reptiles, as well as rodents and insects that live in close contact with the cages of the animals of the zoo under study; finding the serotypes Sandiego, Oranienburg, Weltevreden, Braenderup and Derby. The intraserotype analysis using PFGE showed indistinguishable strains (no genetic diversity), with some isolated exception, which suggests the circulation of this variant of Salmonella among the animal inhabitants in these environments, which can be disseminated through water, food, fomites or vectors like waterfowl and rodents.
Humans
Humans acquire Salmonella infection after ingestion of contaminated food or water. The disease is mainly caused by host-specific serotypes such as Typhi and Paratyphi, however infections in humans are not restricted to them alone.
Since epidemiological surveillance in Mexico has been documented, typhoid fever has been increasing while paratyphoid fever and other salmonellosis have remained in a constant range of about 100,000 cases per year in the last decade (DGE, 2017a) (Figure 1 and 2), highlighting that these data could be higher if all the events of this condition were recorded.
In 1972 and early 1973 an epidemic of typhoid fever (caused by Salmonella Typhi) occurred in Mexico City, which rapidly expanded to surrounding areas affecting more than 10,000 people (Bessudo et al., 1973; Olarte & Galindo, 1973, González-Cortes et al., 1974). Two other important outbreaks related to the common consumption of contaminated food were reported; the first occurred within a hospital, involving 155 people infected by Salmonella Enteritidis (Chávez-de la Peña et al., 2001); the second took place inside a prison, where 150 inmates presented diarrhea and other symptoms of enteric disease caused by Salmonella Oranienburg (Vázquez-Garcidueñas et al., 2014). In both cases the source of infection was not identified.
Although foods of animal origin have been described as the main cause of Salmonella outbreaks, the presence of this bacterium in healthy and sick individuals can convert the human as a carrier, disseminator and therefore responsible for the transmission of this disease (Zaidi et al., 2006; Hernández et al., 2007; Paniagua-Contreras et al., 2008); this is reflected by placing the human as the third source of Salmonella isolation in Mexico, with 21.46 % (Table 1). This problematic accentuates the need to implement continuous and broader surveillance programs to monitor sources of contamination, with the use of more sensitive and faster techniques for the discrimination of pathogens.
Conclusions
Salmonella is a pathogen widely distributed in Mexico. The phenotypic diversity indicates the presence of at least 216 serotypes circulating in the country, being the serotypes Enteritidis, Typhimurium, Anatum, Agona and Meleagridis the most prevalent. The infectious and resistance capacity of circulating serotypes in Mexico depends on the selective pressure of environmental variations including temperature and rainfall. In the same way, this selective pressure is visible in the exacerbated increase of Salmonella serotypes with multiresistance to antibiotics, which provides the bacteria with one increased virulence factor.
In this review of the last 50 years of Salmonella in Mexico, studies addressing issues related to the interactions of this bacterium with the environment in terms of epidemiology, diversity and resistance were included; however, the information is limited. This compilation benchmarks to strengthen and carry out more research, deepening in topics that allow to elucidate more accurately the role that climatic variations, antimicrobial and environmental resistance (biofilms) play in the adaptation and appearance of more virulent strains of this pathogen, based on genetic diversity through the analysis of genomic data that can relate the dynamics of dispersion, seasonality and traceability. This information will contribute to the application of the corresponding actions by the pertinent authorities for this pathogen control, and thereby allow the reduction of morbidity and mortality caused by Salmonella in the Mexican population.











 texto en
texto en 


