The Pepper huastecto yellow vein virus (PHYVV) is one of the main viruses affecting pepper (Capsicum spp.) plants and other Solanaceae in Mexico (García-Nería & Rivera-Bustamante 2011). The virus is widely disseminated in Mexico, the South of the USA, and Guatemala (Torres-Pacheco et al. 1996, Nakhla et al. 2005). PHYVV is a member of the genus Begomovirus (Subgroup III) and belongs to the family Geminiviridae (Palmer & Rybicky 1997). This virus has as hosts several dicotyledoneous plants, like pepper (Capsicum spp.), tomato (Solanum lycopersicum L.), tobacco (Nicotiana spp.), husk tomato (Physalis spp.), Solanum rostratum D., Cucurbita spp., Helianthus annuus L., Datura spp., Carica papaya L., Sorghum halepense P., and Melia azedarach L. (Garzón-Tiznado et al. 2002). It bipartite genome is transmitted by Bemisia tabaci G, which is widely distributed worldwide and causes the most devastating Geminivirus diseases (Morales & Anderson 2001, Varma & Malathi 2003). The main symptoms of PHYVV in pepper plants are veins yellowing, leaves distortion, yellow mosaic, curling leaves, stunted plants and reduction of yields (Garzón-Tiznado et al. 1993).
Management of this Begomovirus has been based mainly on the chemical control through the use of insecticides against vector insects. This method has resulted particularly effective, costly, and represents a biohazard (Borah & Dasgupta 2012). An effective alternative, without bio-risk, and accepted for the integrated management of Begomovirus is the development of resistant genotypes to this group of pathogens (Shankarappa et al. 2008). The first step for the development of resistant cultivars to diseases is the screening of wild and/or domesticated genetic resources, to be used afterwards in the genetic breeding programs of agricultural crops (Pickersgill 1997). There are several studies that showed that wild relatives of Capsicum maintain high levels of genetic variation (Hernández-Verdugo et al. 2001a, Oyama et al. 2006, González-Jara et al. 2011, Pacheco-Olvera et al. 2012), therefore they can be an important source in the search of resistance to PHYVV. There are several reports of resistance sources to PHYVV in Capsicum. Trujillo-Aguirre & Díaz-Plaza (1995) found genetic resistance to PHYVV and PepGMV in wild populations of Capsicum chinense from Southeast Mexico and Hernández-Verdugo et al. (2001b) and Retes-Manjarrez et al. (2016) found genetic resistance to PHYVV in wild populations of Capsicum annuum from Northwest Mexico. Despite that these resistance sources are promising for the genetic breeding programs of Capsicum, to this date no pepper cultivars resistant to PHYVV have been described yet. This could be due to the lack of studies on the genetic basis of the resistance trait or of new sources of resistance to PHYVV.
Genes that code for germin-like proteins that confer resistance to viruses and bacteria in species like Capsicum spp., Beta vulgaris, Triticum spp., and Hordeum vulgare have been reported (Park et al. 2004, Knetch et al. 2010). In Capsicum spp., Barrera-Pacheco et al. (2008) identified a resistance gene in plants of Capsicum chinense Jacq accession BG-3821, and reported it as resistant to PHYVV and PepGMV, after that León-Galván et al. (2011) naming it as “CchGLP”. Several studies have shown that this gene codes for a germin-like protein, which has been associated to provide an important resistance to PHYVV in the BG-3821 genotype of Capsicum chinense (Anaya-López et al. 2005; León-Galván et al. 2011; Guevara-Olvera et al. 2012, Mejía-Teniente et al. 2015).
It is known that from an agronomic trait to be subjected to selection in genetic breeding programs, it needs to have a genetic basis and must be heritable (Falconer & Mackay 1996). Heritability (h2) measures the proportion of total phenotypical variance due to additive genetic causes, determines the rhythm at which the population mean evolves in response to natural or artificial selection. The most important aspect of h2 is its predictive function in traits of interest to help improve the design of genetic breeding programs. Determination of h2 is one of the main objectives of the genetic study in a metric trait (Falconer & Mackay 1996). Retes-Manjarrez et al. (2016) reported that the PHYVV-resistance trait showed an h2 average of 0.17 in one analyzed generation of three wild populations of Capsicum annuum from Northwest Mexico. However, it is important to continue applying selection on the resistant genotypes to PHYVV to analyze and measure the response to the selection of the resistant trait in two more generations of these genotypes to get a better understanding of the heritability and behavior of this resistant trait to design the best genetic model of introgression of this trait into desirable pepper background.
This study is a continuation of the research done by Retes-Manjarrez et al. (2016), where they found three promising resistant population of wild Capsicum annuum to PHYVV and where the resistant plants generation S0 were self-pollinated individually to get and analyze the resistance and the heritability of the resistant trait to PHYVV in the S1 generation of the resistant plants. On this study we analyze the resistance and heritability of the resistant trait in the S2 and S3 generation of the same resistant lines developed by Retes-Manjarrez et al. (2016). The main objectives of this study were to analyze the behavior of the resistant trait to PHYVV in three lines of C. annuum previously chosen as resistant by Retes-Manjarrez et al. (2016), derived from wild populations of Northwest Mexico, and to estimate the heritability of this trait during two more generations. This will help breeders to count upon a greater diversity of resistance sources, thereby, be able to design better strategies for the incorporation of this PHYVV-resistance trait in future pepper cultivars.
Materials and methods
The source of viral inoculum, the insects vectors, the methodology used to inoculate and to evaluate the resistance were provided and following as Retes-Manjarrez et al. (2016).
Vegetable material. In this study, the lines UAS10, UAS12, and UAS13 S2 generation were used for the first resistant assay of this research. These lines were derived from individual plants first generation S1 previously selected as resistant to PHYVV by Retes-Manjarrez et al. (2016). These lines come from three different wild populations of Capsicum annuum of Northwest Mexico. As susceptible control to PHYVV, the C. annuum cultivar Maverick (United Genetics) was used. Seeds were germinated in trays with 200 polystyrene wells in a germination chamber at 30 ± 2 °C. All experiments were performed with 50-day-old plants under nursery conditions with temperatures between 22 and 34 °C during the whole duration of the study.
Source of viral inoculum. We used the strain PHYVV “M53”, which come from pepper plants Jalapeño type Cultivar Grande (Seminis). The inoculum-source plants were maintained in wooden entomologic cages, insect proof (40 cm length, 40 cm width by 60 cm height) and covered with organza fabric. Inoculating healthy plants of the Maverick cultivar through grafting increased the number of inoculum-source plants.
Identification of the virus. For PHYVV identification, the polymerase chain reaction (PCR) method was used with primers 241F and 241R that amplify a 350 bp fragment from the intergenic region of component A of the virus. PCR analysis used for PHYVV detection followed the description by Torres-Pacheco et al. (1996).
Source and maintenance of the vector insect. We used the insect source Bemisia tabaci biotype B free of the virus. The virus-free white flies were placed on cotton (Gossypium hirsutum L.) plants in wooden entomologic cages covered with organza fabric and kept in a nursery for 6 months at an average temperature of 28 ± 2 °C to obtain enough populations to be able to perform the PHYVV-inoculation assays. Cotton plants were substituted by young plants every two months.
Resistance assays. In this study, we performed two PHYVV-genetic resistant assays. The first assay was performed in February 2013. The insect inoculation and grafting methods were used. The insect inoculation consists of placing a plastic bottle with 20 viruliferous insects at adult stage on individual plants of the different genotypes for a 48 h transmission period. After inoculation, an imidacloprid (Confidor®, Bayer Crop Sciences) treatment was applied to eliminate insects. For the grafting inoculation, the methodology used was the reported by Garzón-Tiznado et al. (1993), which consists in placing individual spikes from the grafted inoculum source plants into the plants to be assessed. We inoculated 46, 34, and 28 plants of lines UAS12, UAS13, and UAS10 (S2) with each method, respectively, plus 100 plants of the Maverick cultivar 50 days after sowing. A completely randomized design was used in which each replicate was of one plant.
The second assay was performed in February 2014. The same methodology of inoculation with insects and grafting was used and the same experimental design applied in the first assay. Insect’s inoculation was performed in 218, 163, and 166 plants generation S3 of the progeny of resistant plants from the first assay of lines UAS12, UAS13, and UAS10, respectively, and 250 plants of the Maverick cultivar. The grafting method was used to inoculate 80 plants of each line and of the susceptible control at 60 days after sowing.
Evaluation of viral resistance. Resistance to PHYVV was assessed based on the level of symptoms, time of symptoms appearance, and viral DNA concentration. Levels of resistance were evaluated through a scale of symptoms from 1 to 9 at 60 days post infection (dpi) in both assays, where 1 is without symptoms and 9 corresponds to a plant with the most severe symptoms, which include dwarf plants, with curling and distorted leaves with a clearly defined mosaic. To determine the time of virus incubation daily readings were made until the first symptoms appeared. Relative quantification of the PHYVV virus concentration was made through real time PCR (qPCR) according to the reactions and conditions described by Carrillo-Tripp et al. (2007). We analyzed nine randomly chosen plants from each genotype, UAS12, UAS13, UAS10, and Maverick from the first assay at 60 dpi. with PHYVV.
Plants that obtained a score equal or less than 4.0 were chosen as resistant and were self-pollinated to be used in the next assay. All plants from both assays were analyzed for the presence of PHYVV viral DNA by PCR to discard escape plants.
Molecular analysis of PHYVV resistance. We analyzed individual leaves of nine plants of each of the assessed lines in the second assay to verify the possible presence of a DNA sequence similar to the gene CchGLP. Plants were taken according to their high or intermediate resistance from lines UAS12, UAS13, and UAS10 S3, plus two samples of leaves of two individual plants of the Maverick cultivar. To detect a similar gene CchGLP, end point PCR was used according to the reactions and conditions described by León-Galván et al. (2011), with small modifications. Modifications consist of different specific primers that were designed based on accession number DQ677335.2, which contains the complete sequence of the CchGLP gene of Capsicum chinense BG-3821. The specific primers used on this study were CCRVF (forward primer; 5´- TTGGCTACCCTAATCTTGA-3´) and CCRVR (reverse primer; 5´-TCCTTGATGAAGCTACGAT-3´), that amplified a predicted fragment of the gen CchGLP of 569 pb. The PCR products (10?l) were separated by agarose gel electrophoresis, stained with Gel Red, visualized in a UV transilluminator (Gel-Doc 2000, BIORAD). The length of the fragments obtained was compared to the 1kb molecular weight marker DNA (GIBCO BRL).
DNA extraction of all analyzed plants was made following Dellaporta et al. (1983) method. PCR products were visualized in 1 % agarose gel. The amplified fragments were purified by commercial microcolumns (PureLink) and, then, sequenced according to Sanger et al. (1977) to confirm the presence of PHYVV and the gene CchGLP. The obtained sequence was compared to other PHYVV and CchGLP sequences registered in GenBank (NCBI). Estimation of sequence similarity of the analyzed sequences was achieved with BLAST (<www.ncbi.nlm.nih.gov/BLAST>; Altschulf et al. 1990).
Data analysis. Data obtained from the assessments of genetic resistance to PHYVV from the two assays and from the data to compare the two inoculation methods were subjected to non-parametric variance analysis with the Kruskal-Wallis test and Dunn median to determine the significance among genotypes (p ≤ 0.05). Data from virus incubation and from DNA viral concentration of PHYVV were analyzed by a parametric variance analysis and through the Tukey test to determine significance among genotypes (p ≤ 0.05). All statistical analyses were performed with the SAS software (SAS 1996).
Heritability. The response to the observed selection leads to an estimation of heritability, in narrow sense also called “realized heritability” (h2), this measures the proportion of the total phenotypical variance that is determined only by the additive genetic variance and, therefore, excludes the contribution due to the dominant and epistatic variance. We estimated h2 by means of the equation h2 = R/S, where R is the response to selection, and S is the selection differential. Response to selection R is determined by the difference between the mean of the progenitor population and the mean of the progeny population. The selection differential (S) was obtained by means of the difference between the mean of the progenitor population and the mean of the plants selected as resistant (Falconer & Mackay 1996).
Results
First genetic resistance assay. Line UAS12 showed a significant higher proportion of resistant plants, less symptoms, longer incubation time, and less amount of viral DNA. No significant differences existed between lines UAS13 and UAS10, but they were significantly different from the Maverick cultivar, under both inoculation methods (insect and grafting) at 60 dpi. (H = 164.95; D.F. = 3, p ≤ 0.0001) (Table 1).
Table 1 Results of the first assay on genetic resistance of lines UAS12, UAS13, UAS10 and the Maverick cultivar inoculated with PHYVV through Bemisia tabaci biotype B and the grafting method. Genotype, inoculation method, incidence of disease: number of resistant plants (NRP) and total tested plants (TTP), average index of symptoms severity (means), range of studied symptoms, time of virus incubation in days, and relative concentration of viral DNA in number of viral copies (qPCR) at 60 dpi.
| Genotype | Inoculation Method | NRP/TTP | Means | Range | Incubation | qPCR | |
|---|---|---|---|---|---|---|---|
| A) | UAS12 | Insects | 38/46 (83 %) | 3.6 c | 1-9 | 29 a | 11.9 c |
| UAS13 | Insects | 10/34 (29 %) | 6.5 b | 2-9 | 14 b | 26.5 b | |
| UAS10 | Insects | 4/28 (14 %) | 7.0 b | 2-9 | 16 b | 35.9 b | |
| Maverick | Insects | 0/50 (0 %) | 8.6 a | 7-9 | 7 c | 73.1 a | |
| B) | UAS12 | Grafting | 35/46 (76 %) | 3.4 c | 1-8 | 29 a | 17.0 c |
| UAS13 | Grafting | 5/34 (15 %) | 7.2 b | 3-9 | 13 b | 36.5 b | |
| UAS10 | Grafting | 4/28 (14 %) | 7.3 b | 1-9 | 15 b | 48.9 b | |
| Maverick | Grafting | 0/50 (0 %) | 8.8 a | 7-9 | 8 c | 93.1 a |
Means comparison made with the Fisher test (p ≤ 0.05). Means with the same letter within each inoculation method indicate non-significant differences.
Line UAS12 showed 38 of 46 (83 %) and 35 of 46 (76 %) resistant plants under inoculation with insects and grafting, respectively (Table 1A). Lines UAS13 and UAS10 had 10 of 34 (29.4 %) and 4 of 28 (14.3 %) resistant plants under inoculation with insects, respectively (Table 1A), whereas under grafting inoculation, there were 5 of 34 (14.7 %) and 4 of 28 (14.3 %) resistant plants, respectively (Table 1B). Levels of average symptoms of lines UAS12, UAS13, and UAS10 were of 3.6, 6.5, and 7.0, respectively, under insect’s inoculation; under grafting, the values were 3.4, 7.2, and 7.3, respectively.
The three lines showed a high variation in the symptoms induced by PHYVV under both inoculation methods. Line UAS12 showed a range of 1 to 9 and of 1 to 8, UAS10 showed 2 to 9 and 1 to 9, whereas line UAS13 showed a range of 2 to 9 and 3-9 under insects and grafting inoculation, respectively (Table 1).
The time for the appearance of the first symptoms was in average of 29, 14, and 16 days for lines UAS12, UAS13, and UAS10, respectively, under insect’s inoculation. With grafting inoculation, the first symptoms appeared in average at days 29, 13, and 15 days for lines UAS12, UAS13, and UAS10, respectively (Table 1).
Viral DNA concentration was on average 11.9 relative copies in line UAS12, whereas for lines UAS13 and UAS10, concentrations were 26.5 and 35.9, respectively, under insect’s inoculation. With grafting inoculation, values were 17.0, 36.5, and 48.9 relative copies in lines UAS12, UAS13, and UAS10, respectively (Table 1).
The Maverick cultivar did not show any resistant plant, and showed an average level of symptoms of 8.6 and 8.8 under insect and grafting inoculation, respectively. The first symptoms appeared on average between days seven and eight under both inoculation methods; viral concentration was of 73.1 and 93.1 viral copies under insects and grafting inoculation, respectively (Table 1 A-B).
No significant differences existed between the insects and grafting inoculation methods in levels of symptoms, viral DNA quantification, and incubation of the virus of the assessed genotypes at 60 dpi (H = 16.64; D.F. =1, p = 0.956).
The time of appearance of the first symptoms and the viral DNA concentration of PHYVV are negatively and significantly correlated with the average values of symptoms in the assessed genotypes (r = -0.952; p = 0.0001).
Of the plants classified as resistant from this assay, we chose 73 plants of lines UAS12, 15 of line UAS13, and 8 of line UAS10 as resistant progenitor plants. All these chosen plants were self-pollinated to be used in the second assay with the S3 generation of these plants.
Distribution of symptoms of the first assay. A bimodal tendency was observed in two groups of genes in the distribution of symptoms of the three lines (UAS12, UAS13, and UAS10) under the two inoculation methods (Figure 1A).
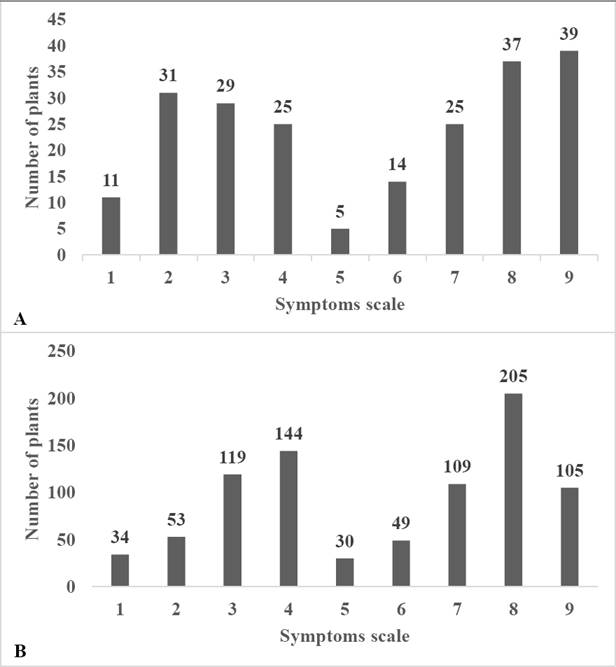
Figure 1 Frequencies distribution of disease symptoms (1-9) of the first (A = S2 generation) and second (B = S3 generation) assays in lines UAS12, UAS13 and UAS10 inoculated with the insects and grafting methods.
Second assay of genetic resistance. Line UAS12 showed a significantly higher proportion of resistant plants, less symptoms in plants, lower incubation time, and less amount of viral DNA. No significant differences existed between lines UAS13 and UAS10, but they were significantly different from the Maverick cultivar under both inoculation methods at 60 dpi. (H = 702.44; D.F. = 3 p ≤ 0.0001) (Table 2).
Table 2 Results of the second assay on genetic resistance of lines UAS12, UAS13, UAS10 and the Maverick cultivar inoculated with PHYVV through Bemisia tabaci biotype B and the grafting method. Genotype, inoculation method, incidence of disease: number of resistant plants (NRP) and total tested plants (TTP), average index of symptoms severity (means), range of studied symptoms, time of virus incubation in days, and relative concentration of viral DNA in number of viral copies (qPCR) at 60 dpi.
| Genotype | Inoculation method | NRP/TTP | Means | Range | Incubation | |
|---|---|---|---|---|---|---|
| A) | UAS12 | Insects | 189/218 (87 %) | 3.4 c | 1-9 | 30 a |
| UAS13 | Insects | 34/163 (21 %) | 6.7 b | 1-9 | 13 b | |
| UAS10 | Insects | 24/166 (15 %) | 7.1 b | 2-9 | 16 b | |
| Maverick | Insects | 0/250 (0 %) | 8.7 a | 7-9 | 8 c | |
| B) | UAS12 | Grafting | 70/80 (88 %) | 3.3 c | 1-9 | 28 a |
| UAS13 | Grafting | 14/80 (16 %) | 7.1 b | 3-9 | 14 b | |
| UAS10 | Grafting | 16/80 (20 %) | 7.1 b | 2-9 | 17 b | |
| Maverick | Grafting | 0/80 (0 %) | 8.8 a | 7-9 | 8 c |
Means comparison made with the Fisher test (p ≤ 0.05). Means with the same letter within each inoculation method indicate non-significant differences.
Line UAS12 showed 189 of 218 (87 %) and 70 of 80 (87.5 %) resistant plants under the insects and grafting inoculation, respectively. Lines UAS13 and UAS10 had 34 of 163 (20.9 %) and 24 of 166 (14.5 %) resistant plants under the insects inoculation, whereas grafting inoculation yielded 14 of 80 (15.5 %) and 16 of 80 (20.0 %) resistant plants, respectively. Under insect’s inoculation, the levels of average symptoms in lines UAS12, UAS13, and UAS10 were of 3.4, 6.7, and 7.1, respectively. With the grafting inoculation, lines UAS12, UAS13, and UAS10 depicted values of 3.3, 7.1, and 7.1, respectively.
The three lines showed again a high variation on the PHYVV symptoms under both inoculation methods. Line UAS12 showed a range of 1 to 9 and line UAS10 of 2 to 9 under both methods, whereas line UAS13 showed a range of 1 to 9 and of 3 to 9 under insects and grafting inoculations, respectively (Table 2).
Appearance of the first symptoms was in average at days 30, 13, and 16 in lines UAS12, UAS13, and UAS10, respectively, under insects inoculation, and of 28, 14, and 17 days, under grafting inoculation, respectively (Table 2).
The Maverick cultivar showed no resistant plants, had an average level of symptoms of 8.7 and 8.8 under insects and grafting inoculation, respectively. The first symptoms appeared in average at 8 days under both inoculation methods (Table 2).
There were no significant differences between the inoculation methods in the levels of symptoms, and in the incubation of the virus of the assessed genotypes at 60 dpi (H = 2.60; D.F. = 1, p = 0.1064).
The time of appearance of the first PHYVV symptoms was negatively and significantly correlated to the average values of the symptoms in the assessed genotypes (r = -0.791; p = 0.0001). Both methods had an infection efficacy of 100 % as viral DNA of PHYVV was detected in all plants of both assays by PCR method.
Distribution of symptoms in the second assay. Again, a bimodal tendency was observed of two groups of genes in the general distribution of symptoms of the three assessed lines under both inoculation methods (Figure 1B).
Molecular analysis from resistance Lines. The amplification of the expected 569 bp fragment was successfully detected in the 27 plant samples of the resistant lines UAS12, UAS13 and UAS10 from the second assay (Figure 2). The sequencing analysis of the amplified fragment from the line UAS12, revealed a 98 % identity with accession DQ677335 that corresponds to the gene CchGLP recorded in the GenBank database (León-Galván et al. 2011).
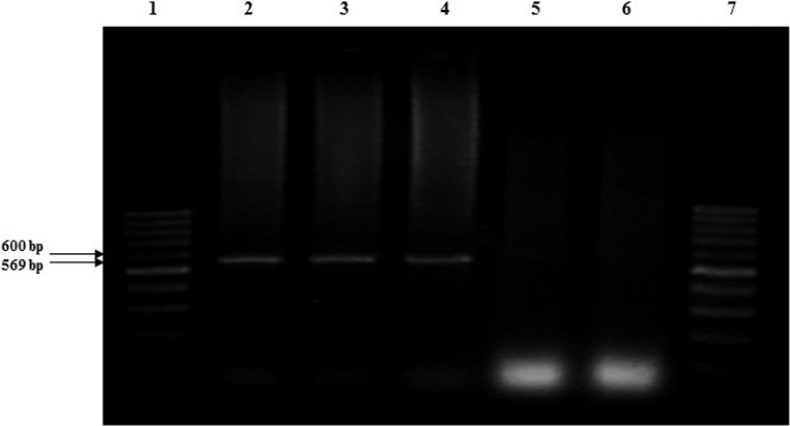
Figure 2 Detection of the predicted 569 bp fragment of one part of the gene CchGLP with pair of primers: CCRVF/CCRVR designed based on accession number DQ677335.2 deposited in the GenBank by León-Galván et al. (2011). Lane 1 and 7, MTM 1kb Plus DNA Ladder. Lane 2, 3 and 4, 569 bp predicted fragment amplified of the DNA extracted from one resistant plant of each line UAS12, UAS13 and UAS10. Lane 5 and 6, samples from the susceptible control (Maverick cv.).
Heritability to PHYVV resistance. The h2 of PHYVV-resistance of line UAS12 was of 0.35 under insect’s inoculation and of 0.58 under grafting inoculation in the first assay (Table 3 A-B). In the second assay, line UAS12 showed an h2 of 0.26 under insect’s inoculation and of 0.10 under grafting (Table 3C-D). Lines UAS13 and UAS10 showed an h2 of zero or close to zero in the first and second assays for the resistance trait under both inoculation methods (Table 3).
Table 3 Heritability values for the resistance to PHYVV in wild genotypes of pepper plants. Inoculation method, genotype and number of generation, means of symptoms of base population, mean of plants selected as resistant, mean of the progeny, response to the selection (R), selection differential (S), and heritability (h2).
| Inoculation method | Genotype/Nº generation | Mean of the base population | Mean of selected plants | Mean of the progeny | R | S | h2 | |
|---|---|---|---|---|---|---|---|---|
| A) | Insects | UAS12/3 | 4.0 | 2.9 | 3.6 | 0.4 | 1.0 | 0.35 |
| UAS13/3 | 6.4 | 4.0 | 6.5 | -0.1 | 2.4 | 0.00 | ||
| UAS10/3 | 6.2 | 3.0 | 7.0 | -0.7 | 3.2 | 0.00 | ||
| B) Grafting | UAS12/3 | 4.0 | 2.9 | 3.4 | 0.6 | 1.1 | 0.58 | |
| UAS13/3 | 6.4 | 4.0 | 7.2 | -0.8 | 2.4 | 0.00 | ||
| UAS10/3 | 6.2 | 3.0 | 7.3 | -1.0 | 3.2 | 0.00 | ||
| C) Insects | UAS12/4 | 3.6 | 2.7 | 3.4 | 0.2 | 0.9 | 0.26 | |
| UAS13/4 | 6.5 | 3.4 | 6.7 | -0.3 | 3.1 | 0.00 | ||
| UAS10/4 | 7.0 | 3.0 | 7.1 | -0.1 | 4.0 | 0.00 | ||
| D) Grafting | UAS12/4 | 3.4 | 2.4 | 3.3 | 0.1 | 0.9 | 0.10 | |
| UAS13/4 | 7.2 | 3.2 | 7.1 | 0.1 | 4.0 | 0.02 | ||
| UAS10/4 | 7.3 | 2.5 | 7.1 | 0.2 | 4.8 | 0.04 | ||
Discussion
Results of this study indicate that line UAS12 showed the highest and significant resistance degree because it presented the highest percentage of resistant plants, lower level of symptoms, longer time for the appearance of the first symptoms, and less relative amount of viral DNA, followed by lines UAS13, UAS10 that showed an intermediate resistance degree, and the Maverick cultivar that showed a high degree of susceptibility in both assays under both inoculation methods (Tables 1 and 2). These results indicate that different PHYVV resistance degrees existed among the assessed lines and, hence, these vary in their level of resistance to PHYVV (Tables 1 and 2). The present results agree with those reported by Hernández-Verdugo et al. (2001b), Godínez-Hernández et al. (2001), and Retes-Manjarrez et al. (2016), who showed different levels of genetic resistance to PHYVV in wild populations of Capsicum in Mexico.
Line UAS12 showed the highest resistance to PHYVV; therefore, it is the most promising to be used in the resistance pepper breeding programs to this virus. On the other hand, lines UAS13 and UAS10 showed an intermediate resistance that can be used to complement the breeding programs as their level of resistance can be regulated by genes different from those of line UAS12 and, therefore, can be useful to improve or make more stable the resistance to PHYVV through genetic breeding methods. These results coincide with those reported by Retes-Manjarrez et al. (2016), who indicated that the UAS12 population showed a higher resistance, whereas UAS13 and UAS10 population showed an intermediate resistance. The present results indicate that the resistance levels of lines UAS12, UAS13, and UAS10 are kept after three generations and that the resistance trait is inherited in a stable way.
The ranges of symptoms among the three lines (Tables 1 and 2) indicate that there is a high variation among them in their PHYVV resistance. This variation in the resistance allowed us to select plants with low indices of symptoms within each line in search of resistance against PHYVV and its show the need to continue with the self-pollination of those plants with the lowest severity indices to obtain more homogeneous lines in terms of their PHYVV-resistance trait to be used in further genetic studies to elucidate the genetic basis of the resistance trait. Further studies to get more homozygous lines are already in progress.
The relative amount of PHYVV viral DNA revealed that line UAS12 had the lowest and significant average value, followed by lines UAS13 and UAS10, which showed a significant difference as compared to the Maverick cultivar. The viral replication in the resistant plants of the lines UAS12, UAS13, and UAS10 inoculated with insects and grafting was less efficient in 84, 64, 51 % and in 82, 61, 48 %, respectively, compared to the replication observed in the susceptible plants of the susceptible control. These results agree with those of García-Nería & Rivera-Bustamante (2011), who reported a viral replication of PepGMV of 70 % lower in accession BG-3821 resistant to PHYVV as compared to its susceptible control. These results also agree with those of Hernández-Verdugo et al. (2001b), Godínez-Hernández et al. (2001), Anaya-López et al. (2003), and García-Nería & Rivera-Bustamante (2011), who reported lower relative concentrations of PHYVV viral DNA among the genotypes considered as resistant and their susceptible controls against this virus in wild populations of Capsicum. The results suggest that lines UAS12, UAS13, and UAS10 possess defense mechanisms that diminish the viral DNA concentration.
The time of appearance of the first symptoms and the PHYVV viral DNA concentration correlated negatively and significantly with the average values of symptoms in the studied genotypes in the two assays under the same inoculation methods. The plants from lines with low levels of symptoms presented a significant delay in the time of appearance and low viral DNA concentration, indicating that the greater the delay in symptoms appearance and the lower the viral DNA concentration, a greater possibility exists that genotypes delay the expression and show low levels of symptoms severity.
According to the distribution of symptoms observed in the two assays, a bimodal tendency of two groups of genes was observed in all lines under the two inoculation methods and in the two assays (Figures 1 and 2). These results suggest that there are at least two genes involved in the resistance to PHYVV in these lines of wild Capsicum annuum L. These agrees with the report by García-Nería & Rivera-Bustamante (2011) who reported that the resistance trait to mixed infections by PHYVV and PepGMV in accession BG-3821 of Capsicum chinense is controlled by two genes with epistatic effects. On the other hand, Retes-Manjarrez et al. (2016) observed the same bimodal tendency and high levels of variation on the symptoms of PHYVV and according to these results they also suggest that this resistant trait most be controlled by at least more than one gene. Unfortunately, there is no more literature about the genetic base of the resistance trait to PHYVV on pepper crop. Further studies with the S4, S5, S6 and S7 generation of these resistant lines are in progress to determinate and elucidate the genetic base of this important trait for future breeding programs.
The amplification of the expected 569 bp fragment from tissue leaf samples of resistant plants of the lines UAS12, UAS13, and UAS10 from the second assay, plus the similarity of 98 % of our sequence with the CchGLP gene (accession DQ677335.2), indicated the possibility of the presence of an open reading frame of PHYVV resistance similar to the reported by León-Galván et al. (2011). The results of this analysis agree in size, sequence, and association with the resistance to PHYVV found by León-Galván et al. (2011), Guevara-Olvera et al. (2012) and Mejía-Teniente et al. (2015).
In the first assay, line UAS12 S2 showed an h2 of 0.35 and 0.58 with mean symptoms of 3.6 and 3.4 under the insects and grafting inoculation methods, respectively. In the second assay, line UAS12 G3 showed an h2 of 0.26 and 0.10 with mean symptoms of 3.4 and 3.3 under the insects and grafting inoculation methods, respectively. These results indicate that line UAS12 showed a response to selection in its S2 and S3 and that the level of resistance presented an increasing pattern of resistance with respect to the S0 and S1 generations, in which Retes-Manjarrez et al. (2016) reported a mean of symptoms of 4.6 and 4.0, respectively, with an h2 of 0.25. On the other side, lines UAS13 (S2 and S3) and UAS10 (S2 and S3) showed an h2 of zero or close to zero without any gain in the levels of resistance in both assays with both inoculation methods in their S2 and S3 generation. The latter indicates that the resistance trait maintains variability in line UAS12, whereas in lines UAS10 and UAS13 the resistance trait has been relatively fixed. The variability maintained by line UAS12 together with the resistance of lines UAS10 and UAS13 could be used to improve this trait. Therefore, the three lines are resistance sources that can be used in the genetic breeding programs. It has been reported that sources with moderate or low resistance to a Begomovirus in tomatoes (Solanum lycopersicum L.) are also useful for breeding programs, because genes that grant moderate or low resistance can complement those of high resistance by pyramiding of these genes in individual genotypes and, thereby, achieving a more stable resistance with less probability of disrupting the resistance due to accumulation of major or minor genes (Hutton et al. 2015).











 nueva página del texto (beta)
nueva página del texto (beta)


