Study contribution
This study describes an alternative, low-cost method that allows farmers to make decisions when selecting animals based on body weight.
Introduction
Image-processing technologies have developed rapidly and can be used to quantitatively characterize the size, shape, and density of organisms or objects.1 Currently, digital image analysis (DIA) is used in many fields, such as human medicine,2 veterinary medicine3 and forensic sciences. 4 Similarly, currently this technology is being used in animal science for carcass characteristics and composition predictions of meat products.5,6 Digital image analysis has been used to estimate body condition, live weight, and carcass composition in heifers,7 beef and dairy cattle,8-14 sheep,15 pigs,16 yaks,17 camels,18 and broiler chickens.19,20
The determination of body weight (BW) is one of the more accurate methods to determine growth and plays an important role in management decisions to support livestock production.7,21 Determination of BW in livestock through image analysis is an emerging research area, allowing the possibility to automatically measure the dimensions of animal images and use prediction equations to establish the relationship between them and live BW.22 The most common approach is the use of images based on the lateral and top areas of the animal that provides different body measurements such as withers height, rump height, body length, diagonal body length, rid depth, chest depth, chest width, thorax width, abdomen width, and dorsal height, that can be correlated with BW.8,22,23
Various techniques have been reported to measure or estimate the BW of livestock as alternatives to the use of weight scales, which is still the most accurate method used by small farms, but it is time-consuming. Also, this is a stressful management situation that in some cases leads to temporal BW losses from animals, and this management includes large fasting periods and deprivation of water derived from holding the sheep in handling pens and these losses can be from 1.8 to 2.9 kg or from 3.5 to 5.6 % BW.21,24
Therefore, it is important to develop alternative and practical methods that are low-cost and easy to implement for decision support systems. Among the alternative methods, the use of biometric measurements (BM) has previously been suggested.24-26 Some authors have reported that manual measurement of BM or body condition score (BCS) of animals is a time-consuming and stressful task for both the farmer and the animal.7,27 Recently the use of DIA has been applied to determine the BM and BW of cattle.8,17,28-30 However, in sheep breeds, the use of DIA to predict their BM, BW, and body size is limited and has not been used in on-farm situations.15 Moreover, it has been reported that in tropical conditions, sheep production systems are characterized by low inputs, and most hair sheep breeds are used. Compared with wool breeds, hair sheep breeds such as Pelibuey are small, with a slow growth rate and poor muscular conformation.31 The objective of this study was to use DIA to predict biometric measurements in Pelibuey sheep as a non-invasive approach under on-farm conditions.
Materials and methods
Ethical statement
Animals were handled according to the guidelines and regulations for animal experimentation of the Department of Agricultural and Livestock Sciences (División Académica de Ciencias Agropecuarias) of the Autonomous Juárez University of Tabasco (Universidad Juárez Autónoma de Tabasco).
Animals, diets, and handling
The data and digital images were collected from 65 nonpregnant and nonlactating Pelibuey ewes aged 2 to 4 years. Animals were selected from a flock composed of 407 adult sheep of the “El Rodeo” farm located at 17° 84’ N latitude and 92° 81’ W longitude, km 14 along the Villahermosa-Jalapa highway in the “Rancheria Víctor Manuel Fernández Manero” in Jalapa, Tabasco, México. The sheep were grouped in pens within a roofed building with a concrete floor and no walls. The diet consisted of a mixed ration of 66 % forage and 34 % concentrate, with an estimated metabolizable energy of 0012 MJ/kg dry matter (DM) and 10 % crude protein (CP)32 The dietary ingredients were cereal grains (corn or sorghum), soybean meal, and hay of tropical grasses, vitamins, and minerals.
Biometric measurement evaluation
Individual biometric measurements were taken before feeding at 08:00, considering the BM described by Bautista-Díaz et al.25 For that, withers height (WH) was measured from the highest point over the scapulae, vertically to the ground. Body length (BL) was measured as the distance between the dorsal point of the scapulae and the ventral point of the tuber coxae. Body diagonal length (BDL) was measured as the distance between the ventral point of the tuber coxae and the cranial point of the shoulder, and finally, rib depth (RD) was measured vertically from the highest point over the scapulae to the endpoint of the rib (at the sternum) (Figure 1). All BMs were recorded in cm. Flexible fiberglass tape and a 65 cm caliper were used for measurements. In addition, the animals were weighed using a digital scale (EQB Model, Torrey, México). Body weight and BM were taken from the animals while standing in a sheep chute.
Digital image analysis
A digital camera with 10.2 megapixels (Sony DSLR-A200, San Diego, USA) was used to take photographs. The camera was set to a standard quality (3872 4000 by 2 592 pixels resolution). The images were taken immediately after the BM was recorded on the same day for all animals. The distance between the digital camera and the chute was the same for all animals, and a metal ruler of 30 cm was fixed on the sheep chute and was used for calibration as described by Ozkaya30 (Figure 1). Three photographs were taken of each animal from the left flank. The best image was selected using the criteria that the entire body of the ewe should stand inside the rectangular area set up and that the image should be of good (clear and steady) resolution as described by Wongsriworaphon et al.33 (Figure 1).
The images were manually measured using ImageJ software ver. 1.51 (https://imagej.nih.gov/ij/index.html) to determine BM calibrated in centimeters. The linear parameters of the lateral profile of WH, BL, BDL, and RD were measured using the tools of the software to DIA as described by Ozkaya.30 The same operator performed the image processing. To orient his shot optimally, the operator placed a mobile cursor at the two ends of each measuring BM. The length of the line was scaled automatically from a pre-programming setup using a 30 cm-long metal ruler for each image.
Statistical Analysis
Descriptive statistical analysis was performed using the MEANS procedure of SAS ver.9.3. Correlation coefficients (r) between variables were estimated using the SAS CORR procedure. The relationships between BM determined by DIA and BM determined in vivo were estimated with linear regression models using the SAS GLM procedure. The precision was assessed by evaluating the r2 of the linear regression of Y (i.e., observed) on X (i.e., predicted).
Also, to assess the predictability of the equations, coefficients of determination (r2), mean square error (MSE), standard deviation (SD), mean square error of prediction (MSEP), and root of the MSEP (RMSEP) were performed and accounted for the distance between predicted values and true values.34 The mean bias (MB), as described by Cochran and Cox,35 was used as a representation of the average inaccuracy of the model, and its calculation is based on the mean difference between observed and model-predicted values.35
The modeling efficiency factor (MEF), which represents the proportion of variation explained by the line Y = X, was used as an indicator of goodness of fit.36,37 The coefficient of model determination (CD) is the ratio of the total variance of observed data to the squared difference between the model-predicted and mean of the observed data. Therefore, the CD statistic explains the proportion of the total variance of the observed values explained by the predicted data.34 The closer to unity the better the model predictions (CD > 1 indicates under prediction and CD < 1 indicates over prediction).
Bias correction factor (Cb), a component of the concordance correlation coefficient (CCC),38 was used as an indicator of deviation from the identity line, and the CCCs were also used as a reproducibility index to account for accuracy and precision. High accuracy and precision were assumed when the coefficients were > 0.80, and low accuracy and precision when the coefficients were < 0.50. Finally, all calculations were obtained using the Model Evaluation System.34
Results and discussion
To our knowledge, the present study is the first to evaluate equations for predicting biometric measurements of Pelibuey sheep as a non-invasive approach under tropical on-farm conditions. After image analysis, data from 10 animals were removed because their images had a low resolution for their correct analysis. This was because when the image was taken, animals were not borne equally on all four feet; they were turned or kneeled. Mean values, SD, and coefficient of variation (CV) of the BM determined in vivo and by DIA are presented in Table 1. The measurements determined in vivo and by DIA were positive and moderately (P < 0.0500) correlated with values of 0.43, 0.66, 0.73, and 0.75 for BL, BDL, WH, and RD, respectively. The digital body measures were highly correlated (> 0.95) between each image labeling.
Table 1 Descriptive statistics of biometric measurements of Pelibuey sheep were determined in vivo and by digital image analysis (n = 55)
| Variable | Description | Mean | SD | CV | Maximum | Minimum |
|---|---|---|---|---|---|---|
| Observed measurements | ||||||
| WH | Observed withers height | 63.81 | 3.98 | 6.23 | 70.00 | 56.00 |
| BL | Observed body length | 48.67 | 8.01 | 16.47 | 55.00 | 46.00 |
| BDL | Observed body diagonal length | 59.60 | 9.29 | 15.59 | 68.00 | 56.00 |
| RD | Observed rib depth | 34.28 | 5.36 | 15.63 | 42.00 | 31.00 |
| DIA measurements | ||||||
| WHDIA | Predicted withers height | 59.05 | 3.48 | 5.90 | 67.65 | 48.39 |
| BLDIA | Predicted body length | 37.84 | 3.37 | 8.90 | 45.47 | 31.81 |
| BDLDIA | Predicted body diagonal length | 54.21 | 4.53 | 8.36 | 65.81 | 44.95 |
| RDDIA | Predicted rib depth | 29.64 | 2.52 | 8.49 | 35.20 | 24.96 |
SD: standard deviation; CV: coefficient of variation; DIA: digital image analysis
The regression equations to predict the BM determined in vivo using those obtained by DIA had an r2 of 0.19 for BL, 0.44 for BDL, 0.54 for WH, and 0.56 for RD (Table 2). Additionally, we explored the relationships between BW and BM obtained by DIA but they were not significant. The results of the predictive equations developed in the present study based on the coefficient of determination, suggest that the use of digital image processing had moderate potential to estimate biometric measurements in hair sheep. Compared with the results of Ozkaya30 who reported that the DIA provided very close measurements and therefore highly related to those determined in vivo in female Holstein calves (r2 > 0.99), our results were quite different.
Table 2 Regression equations to predict biometric measurements of Pelibuey sheep using digital image analysis
| Equations | MSE | RSD | r 2 | P-value | |
|---|---|---|---|---|---|
| 1 | WH (cm) = 14.43(±6.37*) + 0.84(±0.11*** )xWH DIA | 7.46 | 2.73 | 0.54 | < 0.0001 |
| 2 | BL (cm) = 38.85(±3.70***) + 0.29(±0.09**)xBL DIA | 4.32 | 2.08 | 0.19 | 0.0050 |
| 3 | BDL (cm) = 36.70(±4.07**) + 0.44(±0.07**)xBDL DIA | 5.35 | 2.31 | 0.44 | < 0.0001 |
| 4 | RD (cm) = 12.22(±2.80**) + 0.76(±0.09**)xRD DIA | 3.00 | 1.73 | 0.56 | < 0.0001 |
P-value: *P < 0.0500, **P < 0.0100, ***P < 0.001. r2: determination coefficient to predict the biometric measurement; MSE: mean square error; RSD: residual standard deviation. Values in parentheses are the standard errors (SEs) of the parameter estimates. WH: withers height, BL: body length, BDL: body diagonal length and RD: rib depth; DIA: Digital Image Analysis
For the evaluation of the developed equations (Table 3; Figure 2), the equations had low to moderate precision (; Table 3). However, all equations had moderate to high accuracy (bias correction factor > 0.69; Table 3; Figure 2), confirming a moderate reproducibility index and concordance with the observed data (CCC = > 0.60) except the equation for BL (CCC = 0.30). In addition, the positive mean bias (difference between observed and model-predicted values) and the CD value (CD > 1) indicates an under-prediction, suggesting a shift of the model-predicted values to the left of the Y = X line.34 Relating to the MEF, the equations presented a low efficiency of prediction (from 0.13 to 0.55) and indicated a low concordance between the observed and predicted values considering that a perfect fit is equal to 1.
Table 3 Mean and descriptive statistics of the accuracy and precision of the equations for predicting biometric measurements of Pelibuey sheep using digital image analysis (n = 55)
| Variable1 | WH | BL | BDL | RD |
|---|---|---|---|---|
| Mean | 63.4 | 49.3 | 60.5 | 34.6 |
| Standard deviation | 2.89 | 0.97 | 2.00 | 1.91 |
| Maximum | 70.5 | 51.5 | 65.6 | 38.9 |
| Minimum | 54.5 | 47.6 | 56.5 | 34.6 |
| r2 | 0.53 | 0.18 | 0.26 | 0.55 |
| Concordance correlation coefficient | 0.69 | 0.30 | 0.60 | 0.71 |
| Bias correction factor | 0.94 | 0.69 | 0.90 | 0.92 |
| Modeling efficiency | 0.52 | 0.13 | 0.42 | 0.55 |
| Coefficient of model determination | 1.85 | 4.22 | 2.25 | 1.79 |
| Regression analysis | ||||
| Intercept (β0) | ||||
| Estimate | -0.08 | 0.41 | -0.46 | -0.12 |
| SE | 8.24 | 16.56 | 10.29 | 4.31 |
| P-value (β0 = 0) | 0.9973 | 0.9845 | 0.9608 | 0.9767 |
| Slope (β1) | ||||
| Estimate | 1.00 | 1.00 | 1.01 | 1.00 |
| SE | 0.12 | 0.33 | 0.16 | 0.12 |
| P-value (β1 = 1) | 0.9556 | 0.9989 | 0.9300 | 0.9332 |
| MSEP source, % MSEP | ||||
| Mean bias | 2.25 | 5.92 | 2.31 | 1.52 |
| Systematic bias | 0.01 | 0.01 | 0.01 | 0.01 |
| Random error | 97.7 | 94.1 | 97.7 | 98.5 |
| Root MSEP | ||||
| Estimate | 2.71 | 2.09 | 2.29 | 1.71 |
| % of the mean | 4.27 | 4.23 | 3.78 | 4.93 |
1 Obs: observed evaluation data set; MSEP: mean square error of the prediction. WH: withers height, BL: body length, BDL: body diagonal length, and RD: rib depth. r2: determination coefficient for the observed vs. predicted values.
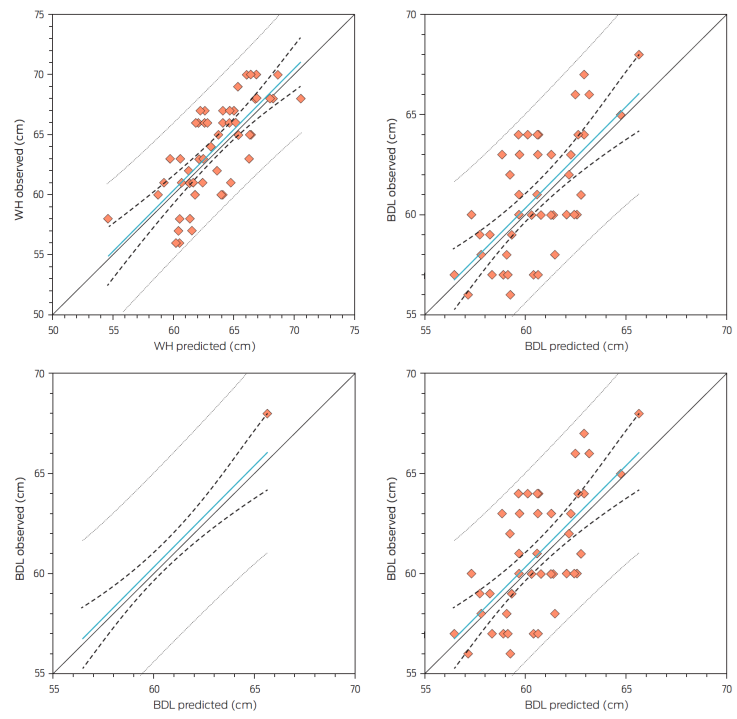
Figure 2 Relationship between the observed and predicted values for predicting biometric measurements of Pelibuey sheep using digital imaging. The solid line is Y = X, and the dotted line is the linear regression. WH: withers height, BL: body length, BDL: body diagonal length, and RD: rib depth. Data was based on 55 animals and one image per animal was used for DIA.
The CD ranged from 1.79 to 4.22, indicating high variability in the predicted data. In this case, a CD value > 1 indicates under-prediction and a CD value < 1 indicates over-prediction (perfect fit = 1).34 In all equations, a small proportion of the error of prediction was associated with the slope, and the main component of MSEP was a random error (> 94.08 %), which indicated small biases in the predictions. In all equations, the RMSEP accounted for 3.78 % to 4.93 % of the observed BM. Moreover, the null hypothesis (intercept = 0 and slope = 1) was accepted for all equations. The prediction model for BL presented the lowest precision (r2 = 0.19), which according to Na Zhang et al.,18 could be due to the change in the animal´s body posture at the time of shooting the photographs. This factor has a great influence on body length and therefore, these authors recommend taking at least five photographs at different times and positions.
In this study, results from the regression equations that were produced had poor predictive capacity. That may be related to potential errors such as: a) an operator error, b) uncertainty in cursor placement during DIA, c) scaling error because of the scale being at one distance and focal length, and the points of measurement being a further distance and the impact of perspective, and d) animal body position between the in vivo measurements and DIA image capture that might lead to different answers from repeated measurements and different postures. In this regard, Khojastehkey et al.18 indicated that the precision of the model is affected by various factors, such as image quality, the type of statistical model, the number of records, the nature of the biometric measurement, and the position and posture of animals. This last factor has been reported in a similar study where live weight in pigs was estimated based on DIA.33 Also, some authors noted that the quality of the prediction of live weight is somewhat sensitive to the environment and the animal’s position33,39 and stated that in field conditions these requirements are not practical and difficult to obtain. The dilemma in this case is that under on-farm conditions, a controlled environment will be difficult but some of the other variables may be easier to modulate.
To improve precision and accuracy, some technical issues need to be considered. Lasfeto and DaudLetik40 reported that to obtain the right animal body dimensions and body weight, it is fundamental to perform quality checks and calibration of cameras used for the analysis. The pixel values of an animal’s image greatly affect the actual values for body weight and body length. On the other hand, the small sample size used in the present study also influenced the poor predictive capacity of the models. Previously, Gomes et al.8 demonstrated that the models to estimate body weight in beef cattle through DIA could have better precision using a greater number of animals (r2 = 0.84 for 35 animals vs r2 = 0.69 for 15 animals).
In addition, Morota et al.22 mentioned that although the use of digital image analysis has great potential for BW estimation in livestock, some challenges still exist such as the automation of image data storage and statistical analysis. As a final point, although the results of the present study were not very promising, they delivered the basis for further developing the method in tropical sheep breeds. Moreover, it is necessary to evaluate the precision and accuracy of these equations using an independent dataset for verifying that they are not population-dependent. To improve equations accuracy, further studies should consider increasing the number of animals as well as several images obtained per animal. Another suggestion is to develop predictive equations for body weight using either the DIA approach or measuring tape.
Conclusions
Overall, the use of digital image analysis was able to predict biometric measurements in Pelibuey ewes with low to moderate precision (r2 > 0.18 ≤ and ≤ r2 0.55) and accuracy (> 0.69). Factors that may affect the accuracy and precision of the relationships of biometric measurements in vivo to digital image analysis should be investigated. Future studies should address the relationships between digital image analysis and the prediction of body weight, body mass index, and body composition in animals from different physiological and production conditions. Also, to improve precision and accuracy, it is necessary to further evaluate that the equations are not population-dependent.











 nueva página del texto (beta)
nueva página del texto (beta)




