Mexico is the leading exporter of Hass avocado (Persea americana) in the world. The fruit is an important source of vitamins, fiber, phenols, and minerals that benefit human health (Rosero et al., 2019). The most valuable nutrients of avocado are the unsaturated fatty acids accumulated in the pulp of the fruit (Rosero et al., 2019). Like other fruits, avocados are a source of protein, fiber, and reducing sugars which are exploited by opportunistic phytopathogens. As a consequence of pathogen attack, fruit quality significantly decreases causing severe economic losses for producers.
Postharvest fungal diseases are one of the main threats to the avocado market. The Colletotrichum, Lasiodiplodia, Sphaceloma, Pseudocercospora, Rhizopus, and Fusarium genera are commonly associated with the development of postharvest diseases in avocado (Perera et al., 2020). However, the potential spectrum of phytopathogenic agents of avocado may vary depending on the geographical region and climatic fluctuations. Puebla is the sixth producer of avocado in Mexico and the northern highlands of this state are tagged as a new area for the production of this valuable fruit. As a new area, the emergence of unknown species with infective potential is latent. In 2021, the avocado producers from the northern highlands of Puebla-Mexico reported an aggressive fruit blotch that caused severe damage and substantial economic losses in harvested avocados. In order to address the problem, this investigation was focused on the identification and isolation of the phytopathogen associated with this disease.
Under open field conditions, the symptoms of diseased fruits were loss of turgor, changes in fruit size, eventual mycelium emergence as well as pulp oxidation (Figure 1A). The incidence of fruit blotch was 56.9% in samples of different ripening stages. For further investigation, 50 diseased fruits (n=50) were harvested from 30 plants grown in 10 different plots during April 2021 in Yaonáhuac, Puebla, Mexico (19° 56′ 55″ N, 97° 26′ 26″ W; 1997 masl). These samples were transferred to the laboratory in plastic bags at 4 °C to be immediately sanitized by immersion in 20% sodium hypochlorite for 20 min and rinsed several times with sterile distilled water in a sterile cabinet (Pérez et al., 2021). Posteriorly, these fruits were dried and placed in sterile humidity chambers (n=30) for 10 days at 28 °C and 70% relative humidity in order to identify the microorganisms associated with avocado rot (Pérez et al., 2021).
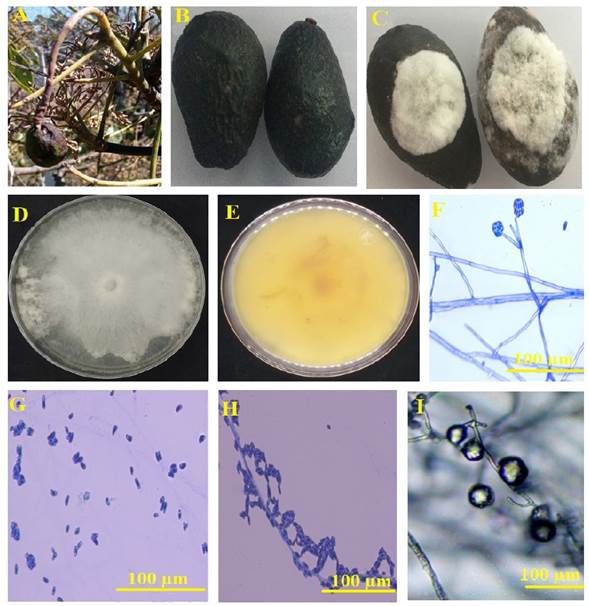
Figure 1 Macroscopic and microscopic features of C. rosea strain C08-9 isolated in the northern highlands of Puebla, Mexico. A, Diseased fruits collected under open field conditions. B, Asymptomatic avocados treated with pure saline solution after 21 days post-inoculation. C, Diseased avocados treated with 1000 conidia dissolved saline solution after 21 days post-inoculation. D, Hyaline mycelium of C. rosea strain C08-9 from PDA cultures of 5 days old. E, Changes in the coloration of PDA medium caused by C. rosea strain C08-9. F, Penicillate conidiophores organized in whorls adhered to hyaline hyphae containing three or four phialides. G-H, Primary conidia showing oval or sub-oval shape. I, Chlamydospores linked to hyaline hyphae
Five different fungal morphologies were observed, isolated, and maintained in potato dextrose agar (PDA; BioxonTM) during these experiments. Preliminary pathogenicity tests were done by inserting hyphal discs (5 mm diameter) of each fungus in healthy avocados (collected in the zone of study) for further incubation in a humidity chamber for 21 days. As a result, one morphology (designed as C08-9 isolate) produced signs of infection on the sixth-day post-inoculation whereas the other fungi did not produce any symptoms after 21 days. Formal pathogenicity tests were carried out in asymptomatic Hass avocados (n=20) using approximately 1000 conidia obtained from a monosporic culture of the isolate C08-9 (10 µL of a sterile saline solution containing 1 × 105 conidia mL-1). The conidia were inoculated by mechanical penetration using a Hamilton syringe in the pericarp of sanitized asymptomatic avocados (n=20) under humidity chamber conditions. The control group (n=20) was only treated with an equivalent volume of saline solution. The microorganism associated with avocado fruit rot was re-isolated and subjected to molecular tests together with the original monosporic culture used. The molecular analyses were done with genomic DNA extracted as previously reported by Pérez et al. (2021). The molecular identity of the associated fungus was obtained by sequencing the internal transcribed spacer (ITS) of the 18S ribosomal gene using the primers ITS1 (TCCGTAGGTGAACCTGCGG) and ITS4 (TCCTCCGCTTATTGATATGC) and the PCR conditions previously reported by Duque-Bautista et al. (2017). The identity was confirmed by amplifying the translation elongation factor 1 alpha gene (TEF-1α) using the primers EF-1H (ATGGGTAAGGAAGACAAGAC) and EF-2T (GGAAGTACCAGTGATCATGTT) (Molnár et al., 2015) using the same PCR conditions described by Duque-Bautista et al. (2017). The corresponding PCR products were purified with the GenElute™ PCR Clean-Up Kit from Sigma-Aldrich Co. (St. Louis Mo. USA) and sequenced at Macrogen Inc. (Seoul, South Korea). The corresponding sequences were stored in the nucleotide database of the National Center for Biotechnology Information (NCBI). The morphological characterization was done with a monosporic culture grown in PDA for 7 days which was exposed to the sunlight for 3 h followed by incubation at 4 °C for 4 h during a period of 10 days with the aim to be stressed. Microcultures prepared with the same medium were subjected to the same stressing process. After 10 days, the cultures and microcultures were subjected to microscopic analysis using a Primo Star Carl Zeiss Primo Star apparatus. The size of the conidia was calculated using the software Carl Zeiss™ AxioVision Rel. 4.8.2. The construction of the phylogenetic tree was done using sequences with high homology to that of C. rosea which were obtained from the NCBI nucleotide database. The sequences of both molecular markers (ITS and TEF-1α) were subjected to multiple alignments using the software Clustal X version 2.0. Posteriorly, the sequences were analyzed using the software MEGA 11.0.11 to be concatenated and to obtain a phylogenetic tree. The sequences subjected to phylogenetic analyses were those of the accessions MK752438.1, MK752494.1, MK752493.1, MK752436.1, MK752439.1, MK752495.1, MK752440.1, MK752496.1, MK752434.1, MK752492.1, MZ433204.1, MZ451398.1, MZ433200.1, MZ451395.1, MZ433199.1, MZ451394.1, MZ433198.1, MZ451393.1, MZ425507.1, MZ451389.1, MT215574.1, MT415234.1, MW199070.1, MW295970.1, OM473286.1 and OM715999.1. Trichoderma stromaticum strain GJS 97-181 was considered as an external parameter to contrast the results. The evolutionary history was inferred using the Neighbor-Joining method. The bootstrap approaches were done with 1000 permutations. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method. There were a total of 2379 positions in the final dataset.
According to the pathogenicity tests, healthy avocados inoculated with sterile saline solution did not present soft rot symptoms after 21 days post-inoculation (Figure 1B). However, avocados inoculated with conidia from the fungus associated with the fruit rot produced evident signs of infection after 6 days post-inoculation but, these were devastating after 21 days (Figure 1C).
The observed symptoms were loss of turgor, changes in epicarp pigmentation (melanin accumulation), and mycelial proliferation which were coincident with the features previously observed in the avocados collected in the zone of study (Figure 1A). The fungus presented white cotton mycelium with fast radial growth in PDA (6.5 mm day-1) and it turned the culture medium to yellow color after 7 days (Figure 1D-1E). The microscopic features of the fungus revealed the presence of penicillate conidiophores organized in whorls containing three or four phialides (Sun et al., 2020) (Figure 1F). The primary conidia showed oval or sub-oval shape (Figure 1G-1H). The average size of the conidia (n=200) was 10.57 µm × 4.42 µm. The presence of chlamydospores was also observed (Figure 1I). These features coincided with the microscopic description reported by Sun et al. (2020) for C. rosea. The partial nucleotide sequences of the 18S ribosomal gene (ITS; 543 bp) and the translation elongation factor 1 alpha gene (TEF-1α; 658 pb) were deposited at the NCBI nucleotide databases with the accessions OM473286 and OM715999, respectively. The ITS sequences kept 99.6% homology with the accession MH864650 whereas the EF-1α sequence had 99.8% homology with the accession KX184998 of C. rosea isolated from soil samples. These results endorsed the identity of the fungus as C. rosea. To the best of our knowledge, there is not any report on the potential pathogenicity of C. rosea in avocados harvested around the world. However, previous investigations describe the devastating effect of a wild strain of C. rosea on Vicia faba, Solanum tuberosum, and Glycine max (Afshari and Hemmati, 2017). The phylogenetic analysis of the C. rosea strain isolated in this investigation (C08-9) revealed that the sum of the branch length was 0.22056366 and the corresponding scale for the concatenated sequences of the ITS-EF-1α markers was 0.020 (Figure 2).

Figure 2 Phylogenetic analysis by the neighbor-joining method generated using MEGA 11.0.11 program with 1000 permutations for ITS/TEF-1 alpha sequences of C. rosea. Trichoderma stromaticum was used as an external reference and the strain C08-9 reported in this study is shown in bold font.
The strain C08-9 of C. rosea isolated in this investigation was grouped within samples reported from diverse countries. The highest homology was with C. rosea isolated from the rotted root from Astragalus membranaceus grown in China, with the strain DG-5-1 isolated from the rotted root of Angelica sinensis, and with the strain Maho4 isolated from wild seeds and fruits collected in Thailand (Perera et al., 2020). To construct a phylogenetic tree, Trichoderma stromaticum GJS 97-181 was considered as an external reference because of its genetic relationship with C. rosea. Our data indicate that C. rosea may have a monophyletic origin and probably the strain C08-9 is coming from a small housing population with pathogenic activity. As is known, the phenotypic variation produced by genetic changes associated with the adaptation process could probably be involved in the development of C. rosea pathogenicity. For further investigation, the strain C08-9 of C. rosea isolated in this investigation was cryopreserved in 15% glycerol at -80 °C in the ceparium of the Dirección de Innovación y Transferencia de Conocimiento at the Benemérita Universidad Autónoma de Puebla, México. As a concluding remark, our investigation suggests the potential pathogenicity of C. rosea in avocado fruit and represents the first step to the generation of subsequent agroecological strategies to control this new threat to the avocado market.











 texto en
texto en 


