1 Introduction
Feature reduction is important in data mining, since its benefits are: simpler and cheaper data collection; less space in memory and disk; smaller processing times, both in training and classification; better visualization and understanding of the results [6].
The reduced set of features should not appreciably decrease the accuracy (percentage of correct answers) of a classifier that uses it.
Moreover, when an object has hundreds of thousands of features, classification becomes complicated, due to ’the course of dimensionality’: as the number of features (dimensions) increase, the data set is very sparsely distributed in the huge spanned dimension space, and many regions of this space are empty [17].
This provokes a curious fact: the distance (for any reasonable metric) between any two points is about the same. Any point is more or less equidistant to all the others [1].
Any three points lie in an almost equilateral triangle. Classifiers that use ’distance’ no longer work well. Such space challenges our intuition about ’closeness’ and ’clustering’, for instance.
Another difficulty arises when data sets are imbalanced, because objects of a certain class are much more abundant than those of other classes.
For instance, sampling the population of a city, the number of people having cancer is quite small compared to the number of people do not having cancer, say, 5% versus 95%.
Many classifiers are tempted to assign to the ’healthy’ class any person, since its error (percentage of incorrect results) will be at most 5%. The main contributions in this paper are:
– FSOC, an algorithm that selects a reduced set of features with less computational effort (much less number of comparisons between features to obtain the subset) than other state-of-the-art feature selection algorithms.
– The classification accuracy when using this reduced set of features is very similar to the accuracy obtained by using the complete set.
– FSOC finds less features than other feature selector algorithms in high dimensional data sets.
– FSOC has also good performance for imbalanced data sets.
The paper is organized as follows. Section 1 introduces the reader to the feature selection area. Section 2, Related works, describes relevant previous work. Section 3 describes the FSOC algorithm and its foundation.
Section 4 compares FSOC with other state-of-the-art selection algorithms, using a statistical analysis. Finally, the last Section contains our conclusions and future work.
The algorithms for feature reduction fall in two groups: feature extraction and feature selection. The first group generates new features by combining the original features.
The number of new features is smaller than the number of the original features. Each object in the dataset is now described by the new features.
The new features are a linear or non-linear combination of the original features, and they are used to span a lower dimensional subspace for the original space, or, in recent subspace techniques, for each pattern class.
Unfortunately, these feature extraction or combination algorithms work mainly with only numeric features or categorical, but not both [14], and they present limitations when the classes are highly imbalanced. That is, when the apriori probabilities of the classes are very different. Therefore, they are not suitable for data sets having a mixture of numerical and categorical features. In addition, the resulting features are difficult to explain to a user that seeks to understand why a particular instance was classified in a certain way.
These techniques reformat, transform and combine the features of each object. For this reason, these algorithms were not considered in this paper.
The second group selects a subset of features from the complete set, called ’relevant features’ because they provide information to correctly discriminate the instances with respect to the class; in other words, the features have a correlation with the class [10, 7].
The goal in both groups is to obtain a good set of features, defined by [9] as those that are correlated with the target class but have little or no correlation with each other. The most important methods for feature selection are filter and wrapper methods.
1.1 Filter Methods
The filter methods carry the process of feature selection without the use of any induction (classification) algorithm. They analyze the training data set to obtain statistical characteristics such as the correlation or the degree of association between two features in order to compare and select features with independence of any predictor (classifier algorithm for instance) and association with the class.
These methods are faster than wrapper methods and generalize better because they act independently of the classification algorithm [15].
1.2 Wrapper Methods
Wrapper methods perform feature selection generating candidate subsets of features, and evaluating them by a previously defined classifier.
Because some inductive algorithm (classifier algorithm for instance) is required, their computational cost is greater than filter methods. In addition, the results will be useful mainly for that classification algorithm [21].
1.3 Ranking Methods
These methods use different correlation measures between the features and the target class, producing ordered lists. The method selects those features that have the highest frequency of appearance in the first places of the lists.
However, the limitation of this type of method is that relationships among features are ignored [11]. In general, the search methods eliminate or add features to the set of relevant features, according to certain selection criteria.
The best-known methods are Forward Selection and Backward Selection.
1.4 Forward Selection
This method starts without any feature in the model (the set of selected features), which implies that no previous information of the correlations between features is necessary.
Every feature that is not included in the current model will be validated through some heuristic.
If it adds discriminatory power to the model, it will be included in it. The method continues until no feature provides information, or if all the features are included in the model.
1.5 Backward Selection
This method is completely contrary to the previous model; it starts with all the features included in the model. Then, it determines first the correlations between any two features for next calculations.
Every feature included in the model is considered for its elimination. It will be excluded from the model if its removal increases the discriminatory power of the model and is redundant with other features. The method continues until no feature can be eliminated.
2 Related Work
Most classifiers in data mining have some weakness when the data set has redundant features or features that are not very relevant. In several publications, metrics have been designed to evaluate the relevance of features [4].
Some metrics only work in numeric, nominal or mixed spaces; for example, the Pearson and Spearman correlation work only with numerical data, while the information gain, the symmetric uncertainty coefficient, V Cramer and confusion measure work with nominal features.
CMCD, based on the theory of class separation, relates numerical and mixed features [13]. However, these metrics only allow measuring the correlation between two features, and do not provide information about whether these features have high correlation with the target class.
Different methods for feature extraction and feature selection has been proposed and used in different areas of knowledge, such as the energy sector, the education sector, recommendation systems, among others.
In article [8], a strategy is presented to increase the efficiency of classifying the stress (low, medium and high) of a driver, through the obtaining and analyzing biological changes such as blood pressure, heartbeat, muscle activity, among others.
The process is carried out through two stages; the first consists of obtaining the new features obtained through feature extraction with the intention of reducing ’altered’ measures and characterizing them.
Subsequently, discriminative common vectors are used to generate an identifying vector for drivers and thus classify them. Moreover, for the discriminative vectors, it is necessary to obtain eigenvectors and eigenvalues to transform the space and it could be limited if the matrices are large.
In addition, the explanation of the final result is confusing, because the vectors generated comes from a series of transformations and combinations between features. In [18] some linear and non-linear techniques for the generation of subspaces are explained, which use Cholesky decomposition to create a matrix that approximates the original data.
The uniqueness of these techniques is that they involve Kernel functions to approximate more complex (non-linear) data. Once this matrix is obtained, the training values can be transformed by its orthogonal representation.
Furthermore, these transformations and new representations of the data cause a lesser understanding of the generated model, and do not always help in decision making. In [2], the use of latent factor models in recommendation systems is proposed because of their ease in dealing with scattered matrices (with missing values).
The main idea is to use the high correlations between columns and rows in order to rotate the axes system and eliminate the redundancy in pair wise correlations. These models map the values of the features into a smaller dimensional space and thereby infer recommendation items based on information from other users.
In [3], Principal Components Analysis (PCA) is used to evaluate and obtain an energy sustainability index for rural communities.
Features such as population density, per capita energy consumption, per capita production, proportion of local residents and tourists, among others, were considered. The result shows that a certain region of France has a better energy index than the rest.
A set of new features is obtained by a linear combination of the original features. Two possible drawbacks are: only numerical features can be combined; and the semantics or meaning of this new set is difficult to understand, and the contribution of the original features is not clear.
Instead, the methods of selecting features maintain the meaning of the original features, since they only exclude less relevant features. In [9], the heuristic called merit is proposed to know how ’good’ a subset of features is.
This heuristic takes into account the usefulness of the features individually, while at the same time it measures the level of correlation between them.
In other words, the merit is the ratio of the correlation between the features with the class divided by the correlation of the features with each other.
The Correlation Feature Selection (CFS) algorithm is developed, its results show great efficiency with different data sets with little or no loss of accuracy in classifiers.
The method allows forward and backward searches. The method consists in adding or eliminating a feature that increases merit and ends when merit decreases while removing or adding a feature, or there are no more features to evaluate.
The main limitation in this method is that although not all combinations of features are generated, too many comparisons are made between features, which could be expensive in a high dimensionality data set. Our proposed FSOC algorithm reduces these comparisons, thus running faster.
[20] describe the method Fast Correlation-Based Filter (FCBF), that seeks primarily the answer to two questions: how to decide if a feature is relevant to discriminate instances with respect to the class (a relevance threshold
In order to answer this question, it uses the condition that the correlation between feature
The method first excludes from the complete set those non-relevant features, and then, every feature in the remaining subset is compared to all others in the subset, to find if there is a strong relationship between any of them.
In this case, it excludes the feature with less discriminatory power. FCBF has two disadvantages. It computes the correlations of the features with the class to provide an acceptable relevance threshold
It excludes features whose correlation with the class are lower than
Another disadvantage is that it considers δ and redundancy sequentially. That is, it first filters out those features that are not relevant (have little discriminating power) and then proceeds to eliminate redundant features.
Although it is a very fast algorithm, the efficiency of the classifiers that use the selected subset of features decreases, which implies loss of discriminating power.
[16] defines four groups of features: (1) strongly relevant features, (2) relevant and non-redundant features, (3) relevant but redundant features and (4) weakly relevant and redundant features.
An optimal subset is one that has features of group 1 and group 2. Their ECMBF algorithm exploits these concepts with two parameters,
The first step is to eliminate features that do not comply with
Moreover, without a previous knowledge of the data set, an initial setting of
In [5] the ANCONE algorithm is developed, that employs the CFS method (Correlation Feature Selection, explained above when describing work [9]), it was used to find personal and socio demographic characteristics associated with the school performance of third grade Mexican High School students in Mathematics.
The complexity of the problem lies in the fact that the data sets to analyze contain approximately 52232 instances (students) and 232 features. Many of these features contain redundant information (Do you have internet at home? Do you have a home computer? Do you have electricity at home?).
From 232 features, 18 were identified as relevant by the CFS method, which are questions about the student’s academic record, the type of school, the educational level of the parents and the student’s academic aspirations. These features increased the efficiency of the classifier from 50% to 68%.
Although the CFS method is effective, when you have a large number of features the algorithm tends to have computationally high costs and requires substantial memory.
Deep learning can be used to extract high-dimensional features, which can be regarded as a complex combination of existing features. This can lead to a reduction of the needed features to accomplish a decent classification.
Therefore, deep learning can be used as a feature reduction method. Nevertheless, it is well known that deep learning takes a long time to converge, especially with massive amount of data having many features. In contrast to this, FSOC is characterized by a short processing time.
3 FSOC Algorithm
The FSOC algorithm uses the heuristic merit
In it,
The same principle is used in test theory to design a composite test for predicting an external variable of interest, ’features’ are individual tests which measure traits related to the variable of interest (class or target feature).
If a group of components increase, it is unlikely that all of them are highly correlated with the target feature and at the same time bear low correlations with each other [19]:
3.1 Description of FSOC
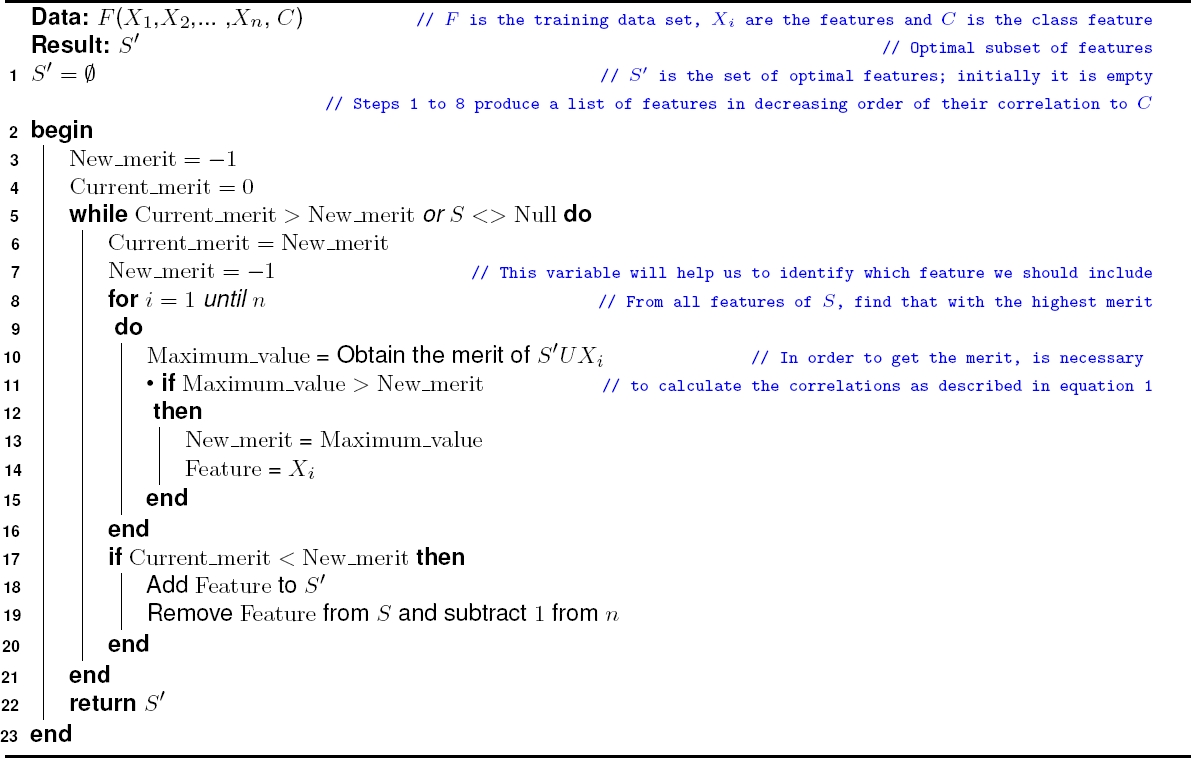
Our algorithm (Algorithm 1, below), shows differences with previous state-of-the-art. FSOC starts by producing a set
Set
Then FSOC evaluates in an orderly manner the inclusion of each element
The feature
The addition of features from
Two important differences between FSOC and CFS algorithms (Algorithm 2) involve the way in which the features to be included in the optimal subset are searched.
The CFS algorithm searches the next feature to be added to the set
If the size of
The first difference is found in steps 4 to 9 of the FSOC algorithm, which obtain the correlation of each feature with the class feature and order them in decreasing order.
In algorithm CFS, its first iteration with the features (steps 7 to 20) also repeatedly seeks the feature
As it turns out, this step (ordering the features) is fundamental to reduce the computational cost. The second and biggest difference appears in steps 13 to 31 of algorithm 1, where two nested cycles are described.
The first cycle adds feature
The second cycle (steps 16 to 26) is internal. Since it searches each feature in decreasing order (of the correlation of the feature with the class), and stops as soon as the next candidate feature
It is considered that the other features (“below”
Instead, CFS evaluates the merits of all the features of
Due to this early stop, FSOC performs fewer comparisons between features, and therefore fewer correlations between them. It is clear that fewer comparisons between features will produce a lower computational cost: lower CPU usage.
With respect to disk I/O, the training set has to be read once into main memory, either all of it at the same time (if it fits) or in batches of objects (if too large to fit in memory), in order to compute the correlations (lines 4 to 8 in algorithm 1; lines 8 to 16 in algorithm 2).
The pseudocode of algorithm CFS (Algorithm 2) does not make clear whether the merit calculation (line 9 in algorithm 2) causes the complete training set to be read for each feature
The end result is that FSOC saves total computational cost = CPU time + I/O time. This improvement is very beneficial for data sets with large volumes of information and with a large number of features.
In addition, because of the way FSOC computes set S’ (algorithm 1), the relevant features in S’ are in ascending order of merit. In this manner, it is easy to reduce further the set of relevant features, in the case S’ is too large.
Now, let us compare how FSOC and ECMBF work, the main difference between FSOC and ECMBF are that ECMBF uses two thresholds;
The search space for these thresholds is two-dimensional in ranges of values in [0-1]; decreasing or increasing them independently does not guarantee that the combination found is ’good’, because the classification accuracy is not necessarily a monotonic function of either of them.
Testing different values of
Now, let us compare how FSOC and FCBF work, the main differences between FSOC and FCBF are that, although both algorithms order the features considering their correlation with the class, the two cycles described in steps 13 to 31 of algorithm 1 allow a faster stop and avoid making comparisons (correlations) between features, as opposed to FCBF, where the search is more exhaustive and therefore considers a greater number of comparisons.
In addition, the relevance parameter (
The next section shows experiments with real data sets where it is observed that FSOC (mainly due to the ordering of features by their correlation with the class and the two nested cycles, already described) helps the reduction in computational costs and number of features selected.
4 Statistical Comparison of FSOC, CFS, ECMBF and FCBF Using Several Classifiers
This section compares FSOC with several feature selection methods, with respect to (1) accuracy (percentage of correct classifications), (2) computational cost (defined as number of necessary comparisons between features), and (3) reduction of features.
The experiments carried out use the accuracy and Kappa measures, since accuracy is an easy measure to understand and although it has a disadvantage when faced with unbalanced data sets, it is complemented by the Kappa measure, that improves on the accuracy measurement by measuring the agreement between predicted and real value, due to chance (the classifier does a random class assignment).
Statistical analysis consists in comparing the algorithms through multiple averages of random executions of data sets.
Our universe U (see Table 1) consists of 36 datasets with nominal, numerical and categorical features, the number of classes ranging from 2 to 26, and the number of instances or objects in the dataset is between 62 and 8,405,0979.
Table 1 Data sets description
| Data set | Area | Instances | # Classes | # Features | Repository | |
| Nominal | Numerical | |||||
| Adult | Social | 32561 | 2 | 8 | 6 | UCI |
| Austra | Financial | 690 | 2 | 8 | 6 | UCI |
| Breast | Health | 683 | 2 | 1 | 9 | UCI |
| Credit | Financial | 653 | 2 | 9 | 6 | UCI |
| Default Credit | Financial | 30000 | 2 | 0 | 23 | UCI |
| Diabetes | Health | 768 | 2 | 0 | 8 | UCI |
| German | Financial | 1000 | 2 | 13 | 7 | UCI |
| Glass | Physical | 214 | 6 | 0 | 9 | UCI |
| Heart | Health | 303 | 2 | 6 | 7 | UCI |
| Iris | Life | 150 | 3 | 0 | 4 | UCI |
| Letter | Recognition | 20000 | 26 | 0 | 16 | UCI |
| Sonar | Physical | 208 | 2 | 0 | 60 | UCI |
| Wine | Chemical | 178 | 3 | 0 | 13 | UCI |
| Cardio | Health | 267 | 2 | 0 | 44 | Keel |
| Coil | Identify | 9822 | 2 | 0 | 85 | Keel |
| Fars | Injury | 100968 | 8 | 24 | 5 | Keel |
| Magic | Physical | 19020 | 2 | 0 | 10 | Keel |
| Ringnorm | Physical | 7400 | 2 | 0 | 20 | Keel |
| Shuttle | Physical | 57999 | 7 | 0 | 9 | Keel |
| Spam | Computer | 4597 | 2 | 0 | 57 | Keel |
| Allaml | Biological | 72 | 2 | 0 | 7128 | scikit-feature |
| Gli_85 | Biological | 85 | 2 | 0 | 22283 | scikit-feature |
| Parkinson | Health | 756 | 2 | 0 | 753 | Kaggle |
| Prostate_ge | Biological | 102 | 2 | 0 | 5966 | scikit-feature |
| Smk_Can | Biological | 187 | 2 | 0 | 19993 | scikit-feature |
| Yale | Face | 165 | 15 | 0 | 1024 | scikit-feature |
| Gisette | Digit | 7000 | 2 | 0 | 5000 | scikit-feature |
| Leukemia | Biological | 72 | 2 | 0 | 7070 | scikit-feature |
| Colon | Biological | 62 | 2 | 0 | 2000 | scikit-feature |
| Madelon | Artificial | 2600 | 2 | 0 | 500 | scikit-feature |
| Pcmac | Text | 1943 | 2 | 0 | 3289 | scikit-feature |
| Basehock | Text | 1993 | 2 | 0 | 4862 | scikit-feature |
| Poker | Game | 1025010 | 10 | 11 | 0 | scikit-feature |
| Susy | Physical | 5000000 | 2 | 0 | 18 | scikit-feature |
| Mobile Health | Health | 1215745 | 13 | 0 | 13 | Kaggle |
| Covid-19 | Health | 8405079 | 4 | 7 | 0 | Kaggle |
To reduce the possible bias introduced by a classifier, three classifiers were used in this analysis: A tree classifier (C4.5), an ensemble of tree classifiers (Random Forest), and a Naive Bayes classifier, in such a way that the averages by the central limit theorem normalize the results and they can be compared each other.
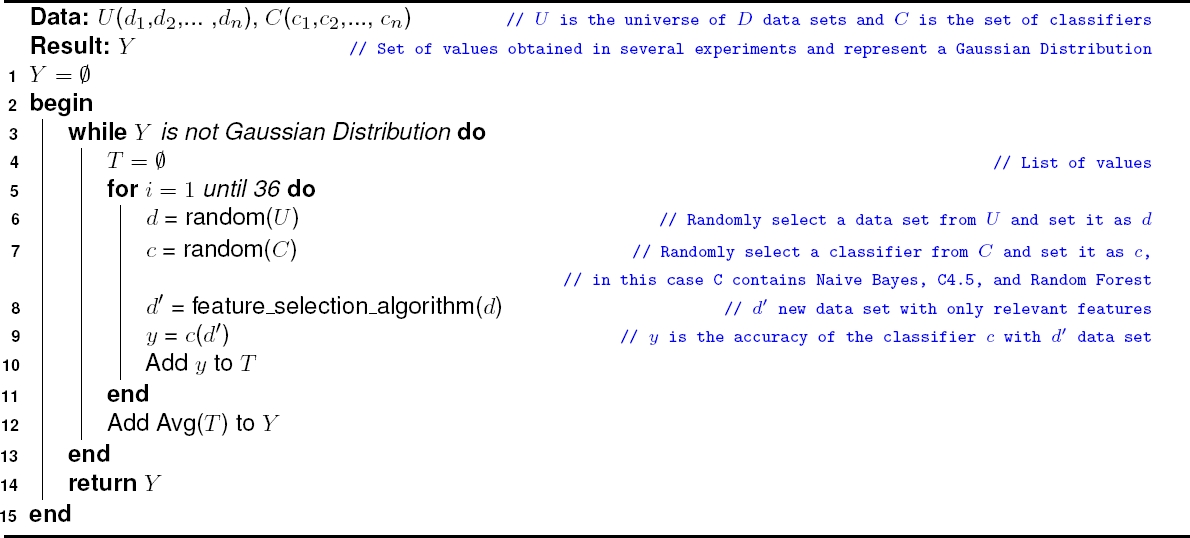
The pseudocode of the statistical algorithm used is shown in algorithm 3. For each feature selection algorithm perform randomly select a data set from
Store the obtained value in
According to the central limit theorem, a Gaussian distribution will be obtained when there are at least 5 observations in each decile and the normality test (a test to determine whether sample data has been drawn from a normally distributed population [within some tolerance]) has an error of 0.05 (This would be equivalent to say that there is a 5% probability that the distribution is not normal)
Comparing the results of FSOC algorithm (Figure 2), when selecting and using the features identified as relevant slightly exceeds the average accuracy than when using the full data set (1.2%), as well as slightly beating the FCBF (3.08%) and ECMBF (1.37%) algorithms.
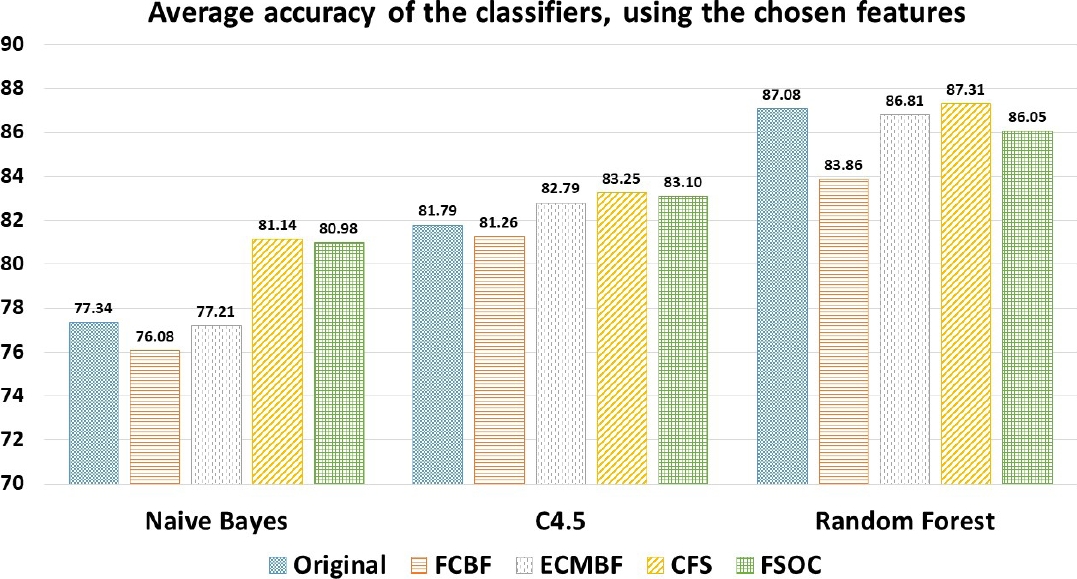
Fig. 1 Average accuracy percentage of the Random Forest, C4.5 and Naive Bayes classifiers for the 36 data sets, using the different features obtained from the feature selection algorithms. The vertical axis of the graph starts at number 68 to allow a greater appreciation of the results. The Feature Selection Ordered by Correlation (FSOC) algorithm has similar accuracy with Correlation Feature Selection (CFS), but with less features and computational cost, Efficient feature selection based on correlation measure (ECMBF) are slightly better just in Random Forest classifier while Fast Correlation-Based Filter (FCBF) is the lowest
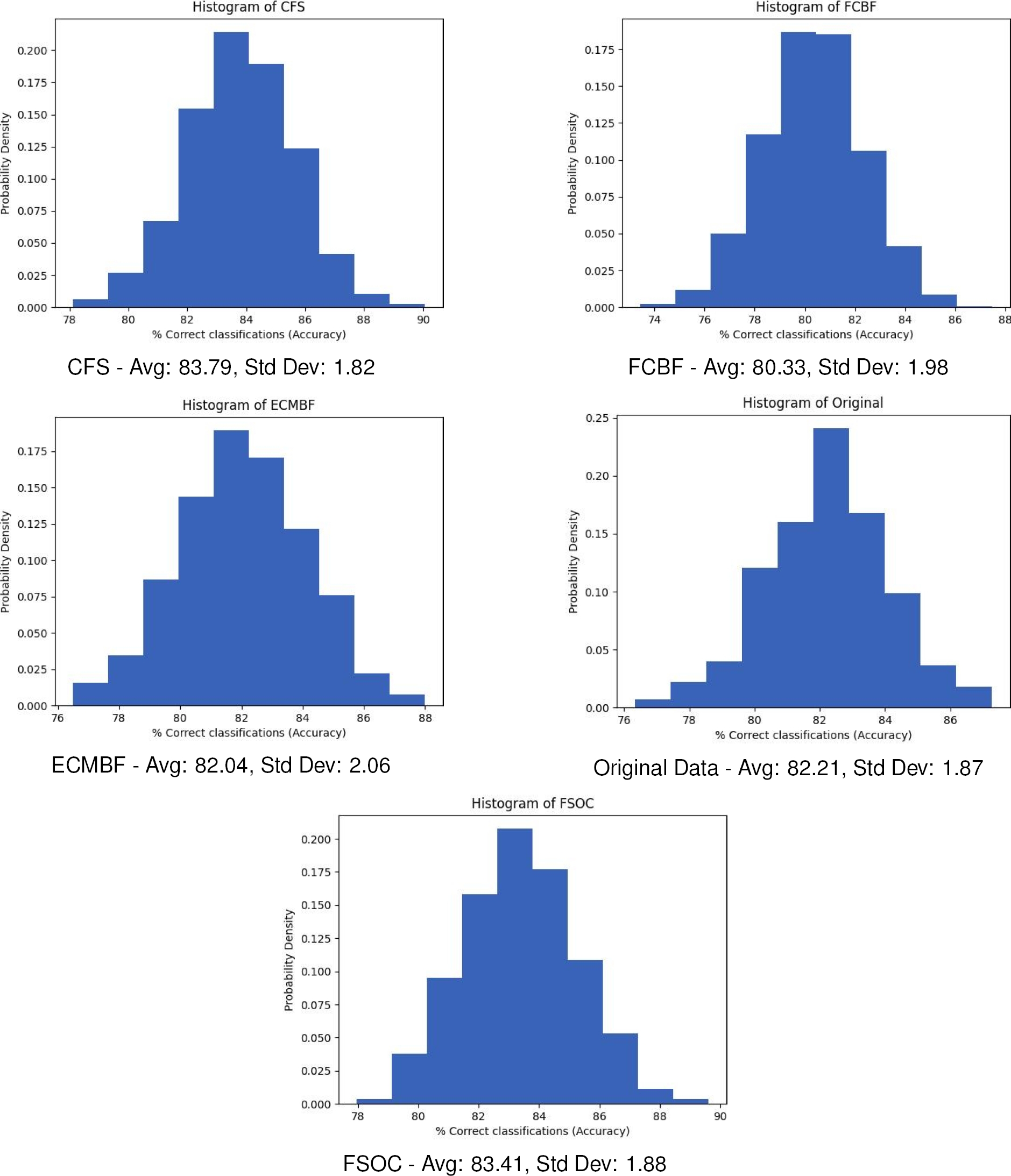
Fig. 2 Results of the statistical algorithm (Algorithm 3) applied to each feature selector algorithm. Gaussian-shaped histograms are displayed, allowing comparisons between them
However, CFS is slightly higher than FSOC (0.38%). The difference of average accuracy between FSOC and the rest of the algorithms it is less than 1%, so we could say that it is practically the same or very similar (see Figure 2).
In addition to the previous results, the Chebyshev inequality [12] (see Equation 2) allows us to rank the algorithms by determining their probability that the percentage of correct classifications (y) is in the interval
If we establish a value of
This is a way of measuring the performance or quality of the algorithm to solve the problems in
Table 2 Quality of each algorithm (last column) given by the Chebyshev inequality in descendent order
| Algorithm | |||
| CFS | 84.20 | 1.89 | 84.20+( |
| FSOC | 83.05 | 1.82 | 83.05+( |
| ECMBF | 82.07 | 1.93 | 82.07+( |
| Original | 81.91 | 1.98 | 81.91+( |
| Data | |||
| FCBF | 80.47 | 2.07 | 80.47+( |
All the algorithms show good average and upper bound in accuracy (greater than 80). While Table 3 shows the algorithms in order from less to high cost computational. Individual results by data set and classifier are placed in Tables 5, 6 and 7.
Table 3 Computational cost, expressed in number of necessary comparisons between features to obtain the optimal subset
| Algorithm | Average | Standard deviation |
| FSOC | 2,236.10 | 798.80 |
| CFS | 97,254.09 | 46,801.22 |
| FCBF | 11,088,895.62 | 6,374,684.55 |
| ECMBF | 13,932,895.10 | 7,728,952.53 |
Table 4 Number of features selected in each feature selector algorithm for each data set and the computational cost required to obtain them. It is observed that FSOC in the vast majority obtains fewer features than the rest of the algorithms with a smaller number of comparisons. The average is rounded to the whole number
| Data set | Original Dara | FCBF | ECMBF | CFS | FSOC | ||||
| #Feat | #Feat | Cost | #Feat | Cost | #Feat | Cost | #Feat | Cost | |
| Adult | 14 | 7 | 44 | 14 | 92 | 5 | 69 | 5 | 34 |
| Austra | 14 | 8 | 42 | 14 | 92 | 1 | 27 | 1 | 16 |
| Breast | 10 | 8 | 31 | 8 | 31 | 9 | 45 | 9 | 45 |
| Credit | 15 | 8 | 54 | 15 | 104 | 1 | 29 | 1 | 17 |
| Default | 23 | 8 | 64 | 22 | 237 | 5 | 123 | 5 | 43 |
| Credit | |||||||||
| Diabetes | 8 | 6 | 22 | 8 | 29 | 3 | 26 | 3 | 17 |
| German | 20 | 9 | 67 | 20 | 191 | 4 | 90 | 4 | 34 |
| Glass | 9 | 8 | 29 | 9 | 37 | 5 | 39 | 5 | 34 |
| Heart | 13 | 8 | 48 | 13 | 53 | 7 | 76 | 7 | 48 |
| Iris | 4 | 3 | 6 | 4 | 7 | 2 | 9 | 2 | 9 |
| Letter | 16 | 12 | 77 | 14 | 118 | 9 | 115 | 9 | 79 |
| Sonar | 60 | 51 | 1263 | 58 | 1563 | 16 | 884 | 16 | 212 |
| Wine | 13 | 11 | 52 | 11 | 60 | 8 | 81 | 6 | 40 |
| Cardio | 44 | 27 | 397 | 41 | 831 | 18 | 665 | 5 | 64 |
| Coil | 85 | 61 | 1918 | 84 | 3492 | 7 | 652 | 6 | 112 |
| Fars | 29 | 8 | 80 | 29 | 407 | 3 | 110 | 3 | 38 |
| Magic | 10 | 3 | 18 | 9 | 46 | 3 | 34 | 3 | 19 |
| Ringnorm | 20 | 20 | 191 | 20 | 191 | 20 | 210 | 20 | 210 |
| Shuttle | 9 | 2 | 16 | 8 | 31 | 3 | 30 | 3 | 27 |
| Spam | 57 | 16 | 358 | 57 | 1597 | 10 | 572 | 10 | 131 |
| Allaml | 7129 | 6168 | 19032070 | 6818 | 23276950 | 28 | 206335 | 9 | 7191 |
| Gli_85 | 22283 | 18882 | 166168130 | 21750 | 236834285 | 53 | 1201851 | 10 | 20045 |
| Parkinson | 753 | 753 | 283881 | 753 | 283881 | 336 | 196728 | 12 | 879 |
| Prostate_ge | 5966 | 5007 | 12546176 | 5264 | 14899715 | 24 | 148850 | 7 | 6007 |
| Smk_Can | 19993 | 18229 | 166168130 | 19428 | 188835632 | 52 | 1058251 | 8 | 20045 |
| Yale | 1024 | 772 | 299532 | 831 | 350156 | 31 | 32272 | 9 | 1086 |
| Gisette | 5000 | 3504 | 6152667 | 4584 | 10678832 | 34 | 174405 | 7 | 5047 |
| Leukemia | 7070 | 6713 | 22538069 | 7066 | 24939737 | 22 | 162357 | 5 | 7094 |
| Colon | 2000 | 1951 | 1904381 | 1946 | 1892599 | 18 | 37829 | 2 | 2005 |
| Madelon | 500 | 500 | 125250 | 500 | 125250 | 9 | 4955 | 3 | 509 |
| Pcmac | 3289 | 3170 | 5027017 | 3253 | 5290982 | 14 | 49230 | 10 | 3354 |
| Basehock | 4862 | 4261 | 9086273 | 4815 | 11608599 | 13 | 67977 | 12 | 4952 |
| Poker | 10 | 10 | 54 | 10 | 54 | 4 | 40 | 5 | 30 |
| Susy | 18 | 18 | 170 | 18 | 170 | 10 | 143 | 4 | 36 |
| Mobile Health | 14 | 14 | 128 | 14 | 128 | 10 | 88 | 10 | 76 |
| COVID-19 | 7 | 7 | 34 | 7 | 34 | 3 | 22 | 1 | 9 |
| Average | 2233 | 1951 | 11370464 | 2153 | 14417394 | 22 | 92922 | 6 | 2211 |
Table 5 Random forest classification for each feature selection algorithm and data set. On Average FSOC is 1.7% approximately below CFS but with less features and computational cost. The results for the SUSY and Poker dataset are incomplete because the model that was built could not be stored in memory
| Data set | Original Data | FCBF | ECMBF | CFS | FSOC | |||||
| %Acc. | Kap. | %Acc. | Kap. | %Acc. | Kap. | %Acc. | Kap. | %Acc. | Kap. | |
| Adult | 84.82 | 0.566 | 82.12 | 0.501 | 84.82 | 0.566 | 85.79 | 0.574 | 85.79 | 0.574 |
| Austra | 86.66 | 0.73 | 83.76 | 0.672 | 86.66 | 0.73 | 85.5 | 0.712 | 85.5 | 0.712 |
| Breast | 97.21 | 0.939 | 97.91 | 0.942 | 97.21 | 0.939 | 97.91 | 0.939 | 97.91 | 0.939 |
| Credit | 86.98 | 0.738 | 85.45 | 0.706 | 86.98 | 0.738 | 86.37 | 0.729 | 86.37 | 0.729 |
| Default_Credit | 81.65 | 0.376 | 79.07 | 0.26 | 86.67 | 0.369 | 81.65 | 0.364 | 81.65 | 0.364 |
| Diabetes | 76.56 | 0.474 | 74.47 | 0.429 | 76.56 | 0.474 | 72.26 | 0.379 | 72.26 | 0.379 |
| German | 75.7 | 0.352 | 72.1 | 0.283 | 75.7 | 0.352 | 69.7 | 0.246 | 69.7 | 0.246 |
| Glass | 79.9 | 0.723 | 78.5 | 0.702 | 79.9 | 0.723 | 74.76 | 0.652 | 74.76 | 0.652 |
| Heart | 81.18 | 0.619 | 79.2 | 0.578 | 81.18 | 0.619 | 79.86 | 0.592 | 79.86 | 0.592 |
| Iris | 95.33 | 0.93 | 95.33 | 0.93 | 95.33 | 0.93 | 94 | 0.91 | 94 | 0.91 |
| Letter | 96.46 | 0.963 | 96.05 | 0.959 | 96.46 | 0.963 | 94.78 | 0.946 | 94.78 | 0.946 |
| Sonar | 86.05 | 0.718 | 83.17 | 0.658 | 87.01 | 0.737 | 83.65 | 0.67 | 83.65 | 0.67 |
| Wine | 98.31 | 0.975 | 97.75 | 0.965 | 98.31 | 0.974 | 97.19 | 0.958 | 97.19 | 0.958 |
| Cardio | 80.14 | 0.233 | 82.39 | 0.308 | 80.89 | 0.286 | 81.27 | 0.339 | 79.02 | 0.259 |
| Coil | 92.86 | 0.077 | 92.5 | 0.06 | 92.79 | 0.073 | 93.5 | 0.044 | 93.95 | 0.035 |
| Fars | 77.77 | 0.698 | 78.38 | 0.704 | 77.77 | 0.698 | 76.68 | 0.68 | 76.68 | 0.68 |
| Magic | 87.98 | 0.728 | 82.51 | 0.602 | 87.68 | 0.721 | 82.51 | 0.602 | 82.51 | 0.602 |
| Ring | 95.29 | 0.906 | 95.06 | 0.901 | 95.29 | 0.906 | 95.29 | 0.906 | 95.29 | 0.906 |
| Shuttle | 99.99 | 0.999 | 94.34 | 0.853 | 99.97 | 0.999 | 99.84 | 0.996 | 99.84 | 0.996 |
| Spam | 95.62 | 0.908 | 94.69 | 0.888 | 95.62 | 0.908 | 92.51 | 0.843 | 92.51 | 0.843 |
| Allaml | 91.66 | 0.805 | 72.22 | 0.294 | 88.88 | 0.735 | 98.61 | 0.969 | 94.4 | 0.875 |
| Gli_85 | 84.7 | 0.591 | 70.58 | 0.079 | 88.23 | 0.696 | 97.64 | 0.944 | 92.94 | 0.83 |
| Parkingson | 85.31 | 0.548 | 87.03 | 0.606 | 85.31 | 0.548 | 87.56 | 0.624 | 86.11 | 0.596 |
| Prostate_ge | 88.23 | 0.764 | 81.37 | 0.626 | 89.21 | 0.784 | 96.07 | 0.921 | 94.11 | 0.882 |
| Smk_can | 68.98 | 0.375 | 66.84 | 0.333 | 59.89 | 0.2 | 81.81 | 0.634 | 73.79 | 0.474 |
| Yale | 77.57 | 0.759 | 75.75 | 0.74 | 74.54 | 0.727 | 75.75 | 0.74 | 64.84 | 0.623 |
| Gissette | 96.9 | 0.938 | 92.77 | 0.855 | 96.62 | 0.928 | 94.88 | 0.896 | 87.45 | 0.749 |
| Leukemia | 93.05 | 0.839 | 80.55 | 0.516 | 93.05 | 0.842 | 98.61 | 0.969 | 97.22 | 0.937 |
| Colon | 85.48 | 0.665 | 77.41 | 0.462 | 80.64 | 0.549 | 87.09 | 0.723 | 88.7 | 0.74 |
| Madelon | 65.84 | 0.316 | 65.65 | 0.313 | 65.84 | 0.316 | 85.57 | 0.711 | 71.76 | 0.435 |
| Pcmac | 94.13 | 0.882 | 91.71 | 0.834 | 94.28 | 0.885 | 86.72 | 0.733 | 85.33 | 0.705 |
| Base | 98.24 | 0.964 | 96.98 | 0.939 | 98.49 | 0.969 | 90.71 | 0.814 | 90.01 | 0.8 |
| Poker | * | * | * | * | * | * | 75.32 | 0.539 | 89.87 | 0.81 |
| Susy | * | * | * | * | * | * | * | * | * | * |
| Mobile Health | 95.11 | 0.91 | 95.11 | 0.91 | 95.11 | 0.91 | 95.54 | 0.905 | 95.54 | 0.905 |
| Covid-19 | 82.24 | 0.585 | 82.24 | 0.585 | 82.24 | 0.585 | 80.93 | 0.555 | 79.15 | 0.53 |
| Average | 87.17 | 0.694 | 84.146 | 0.617 | 86.95 | 0.688 | 87.721 | 0.707 | 86.017 | 0.682 |
Table 6 C4.5 classifications for each feature selection algorithm and data set. On Average FSOC has similar accuracies with respect to CFS, but is better than the complete data set, ECMBF and FCBF
| Data set | Original Data | FCBF | ECMBF | CFS | FSOC | |||||
| %Acc. | Kap. | %Acc. | Kap. | %Acc. | Kap. | %Acc. | Kap. | %Acc. | Kap. | |
| Adult | 85.79 | 0.586 | 85.49 | 0.555 | 85.79 | 0.586 | 85.67 | 0.566 | 85.67 | 0.566 |
| Austra | 86.08 | 0.73 | 84.05 | 0.677 | 86.08 | 0.73 | 85.5 | 0.712 | 85.5 | 0.712 |
| Breast | 96.04 | 0.914 | 95.6 | 0.904 | 96.04 | 0.914 | 96.04 | 0.914 | 96.04 | 0.914 |
| Credit | 85.29 | 0.703 | 86.52 | 0.73 | 85.29 | 0.703 | 86.37 | 0.729 | 86.37 | 0.729 |
| Default_Credit | 80.32 | 0.337 | 79.55 | 0.315 | 86.53 | 0.345 | 82.13 | 0.379 | 82.13 | 0.379 |
| Diabetes | 73.82 | 0.416 | 75 | 0.438 | 73.82 | 0.416 | 74.6 | 0.425 | 74.6 | 0.425 |
| German | 70.7 | 0.25 | 71.5 | 0.266 | 70.7 | 0.25 | 74.6 | 0.25 | 74.6 | 0.25 |
| Glass | 65.88 | 0.541 | 63.55 | 0.492 | 65.88 | 0.541 | 65.88 | 0.652 | 65.88 | 0.652 |
| Heart | 78.54 | 0.567 | 79.86 | 0.591 | 78.54 | 0.567 | 77.55 | 0.544 | 77.55 | 0.547 |
| Iris | 96 | 0.94 | 96 | 0.94 | 96 | 0.94 | 94 | 0.91 | 94 | 0.91 |
| Letter | 87.92 | 0.874 | 84.59 | 0.839 | 87.99 | 0.875 | 87.28 | 0.868 | 87.28 | 0.868 |
| Sonar | 71.15 | 0.422 | 68.75 | 0.369 | 71.15 | 0.422 | 78.84 | 0.574 | 78.84 | 0.574 |
| Wine | 93.82 | 0.906 | 88.76 | 0.829 | 94.38 | 0.915 | 93.82 | 0.906 | 92.13 | 0.88 |
| Cardio | 74.9 | 0.238 | 79.77 | 0.307 | 76.79 | 0.28 | 80.52 | 0.405 | 78.27 | 0.256 |
| Coil | 93.76 | 0.002 | 93.91 | 0.009 | 93.94 | 0.007 | 94.03 | 0.006 | 94.03 | 0.003 |
| Fars | 93.95 | 0.007 | 78.47 | 0.705 | 93.95 | 0.007 | 94.03 | 0.112 | 94.03 | 0.112 |
| Magic | 79.85 | 0.757 | 81.74 | 0.574 | 84.9 | 0.657 | 81.74 | 0.574 | 81.74 | 0.574 |
| Ring | 85.05 | 0.661 | 90.24 | 0.804 | 85.05 | 0.661 | 90.22 | 0.804 | 90.22 | 0.804 |
| Shuttle | 99.97 | 0.999 | 94.7 | 0.862 | 99.95 | 0.999 | 99.81 | 0.995 | 99.81 | 0.995 |
| Spam | 92.93 | 0.852 | 92.16 | 0.835 | 92.93 | 0.852 | 91.84 | 0.827 | 91.84 | 0.827 |
| Allaml | 88.82 | 0.754 | 88.88 | 0.754 | 90.27 | 0.787 | 90.27 | 0.795 | 90.27 | 0.795 |
| Gli_85 | 83.52 | 0.612 | 87.05 | 0.691 | 84.7 | 0.635 | 87.05 | 0.691 | 89.41 | 0.747 |
| Parkingson | 80.95 | 0.466 | 80.95 | 0.464 | 80.95 | 0.466 | 78.96 | 0.398 | 81.61 | 0.476 |
| Prostate_ge | 81.37 | 0.626 | 80.39 | 0.607 | 85.29 | 0.705 | 86.27 | 0.725 | 84.31 | 0.685 |
| Smk_can | 60.42 | 0.195 | 63.63 | 0.261 | 62.56 | 0.238 | 68.98 | 0.378 | 70.58 | 0.408 |
| Yale | 43.63 | 0.396 | 41.81 | 0.376 | 44.24 | 0.402 | 43.63 | 0.396 | 44.84 | 0.409 |
| Gissette | 93.58 | 0.871 | 92.06 | 0.84 | 93.85 | 0.877 | 92.77 | 0.855 | 87.3 | 0.746 |
| Leukemia | 91.66 | 0.816 | 93.05 | 0.842 | 93.05 | 0.842 | 94.44 | 0.875 | 93.05 | 0.842 |
| Colon | 74.19 | 0.386 | 79.03 | 0.506 | 82.25 | 0.573 | 79.03 | 0.517 | 85.48 | 0.658 |
| Madelon | 72.57 | 0.451 | 72.57 | 0.451 | 72.57 | 0.451 | 74.73 | 0.494 | 67.61 | 0.352 |
| Pcmac | 82.55 | 0.65 | 80.95 | 0.618 | 83.47 | 0.669 | 80.64 | 0.611 | 80.64 | 0.611 |
| Base | 91.21 | 0.824 | 87.65 | 0.753 | 91.57 | 0.831 | 86.55 | 0.731 | 86.65 | 0.733 |
| Poker | 64.97 | 0.346 | 64.97 | 0.346 | 64.97 | 0.346 | 73.63 | 0.504 | 75.07 | 0.554 |
| Susy | 79.58 | 0.583 | 79.58 | 0.583 | 79.58 | 0.583 | 78.95 | 0.571 | 78.01 | 0.552 |
| Mobile Health | 91.31 | 0.821 | 91.31 | 0.821 | 91.31 | 0.821 | 91,39 | 0.823 | 91,39 | 0.823 |
| Covid-19 | 82.24 | 0.586 | 82.24 | 0.586 | 82.24 | 0.586 | 80.93 | 0.558 | 79.15 | 0.53 |
| Average | 81.85 | 0.57 | 81.30 | 0.59 | 82.29 | 0.59 | 83.24 | 0.60 | 83.09 | 0.60 |
Table 7 Naive Bayes classification for each data set and feature selection algorithm. On average FSOC is slightly below CFS but with less features and computational costs. Also, FSOC is better than full data set, ECMBF and FCBF algorithm. In the Poker data set, an anomaly is shown, where all the feature selectors and the original set do not present any difference, unlike other classifiers (tables 5 and 6). This is probably due to its high unbalance in the classes and that it is not the best classifier for this set
| Data set | Original Data | FCBF | ECMBF | CFS | FSOC | |||||
| %Acc. | Kap. | %Acc. | Kap. | %Acc. | Kap. | %Acc. | Kap. | %Acc. | Kap. | |
| Adult | 83.47 | 0.501 | 79.79 | 0.323 | 83.47 | 0.501 | 79.93 | 0.327 | 79.93 | 0.327 |
| Austra | 77.53 | 0.531 | 75.5 | 0.484 | 77.53 | 0.531 | 85.5 | 0.712 | 85.5 | 0.712 |
| Breast | 96.04 | 0.914 | 96.48 | 0.924 | 96.33 | 0.921 | 96.33 | 0.921 | 96.33 | 0.921 |
| Credit | 78.25 | 0.55 | 76.87 | 0.52 | 78.25 | 0.55 | 86.37 | 0.729 | 86.37 | 0.729 |
| Default_Credit | 69.35 | 0.289 | 79.63 | 0.324 | 61.83 | 0.22 | 79.53 | 0.356 | 79.53 | 0.356 |
| Diabetes | 76.3 | 0.466 | 76.82 | 0.47 | 76.3 | 0.466 | 76.43 | 0.455 | 76.43 | 0.455 |
| German | 75.4 | 0.381 | 73.5 | 0.314 | 75.4 | 0.381 | 74.1 | 0.319 | 74.1 | 0.319 |
| Glass | 49.53 | 0.334 | 43.92 | 0.239 | 49.53 | 0.334 | 50 | 0.332 | 50 | 0.332 |
| Heart | 82.5 | 0.646 | 84.48 | 0.685 | 82.5 | 0.646 | 83.16 | 0.685 | 82.83 | 0.653 |
| Iris | 96 | 0.94 | 95.33 | 0.93 | 96 | 0.94 | 96 | 0.94 | 96 | 0.94 |
| Letter | 64.01 | 0.626 | 65.49 | 0.641 | 65.81 | 0.644 | 64.63 | 0.632 | 64.63 | 0.632 |
| Sonar | 67.78 | 0.366 | 68.75 | 0.387 | 69.23 | 0.394 | 69.23 | 0.394 | 69.23 | 0.394 |
| Wine | 96.62 | 0.949 | 97.75 | 0.965 | 97.19 | 0.958 | 97.19 | 0.958 | 97.75 | 0.965 |
| Cardio | 68.53 | 0.358 | 67.79 | 0.33 | 67.41 | 0.332 | 70.78 | 0.393 | 72.65 | 0.407 |
| Coil | 78.07 | 0.121 | 87.06 | 0.121 | 77.86 | 0.118 | 93.63 | 0.057 | 93.76 | 0.042 |
| Fars | 77.96 | 0.701 | 78.06 | 0.7 | 77.96 | 0.701 | 76.68 | 0.68 | 76.68 | 0.68 |
| Magic | 72.68 | 0.329 | 76.02 | 0.441 | 73.09 | 0.341 | 76.02 | 0.441 | 76.02 | 0.441 |
| Ring | 97.97 | 0.959 | 97.97 | 0.959 | 97.97 | 0.959 | 97.97 | 0.959 | 97.97 | 0.959 |
| Shuttle | 92.8 | 0.793 | 94.34 | 0.853 | 94.63 | 0.861 | 93.47 | 0.826 | 93.47 | 0.826 |
| Spam | 79.68 | 0.604 | 76.07 | 0.542 | 79.68 | 0.604 | 86.9 | 0.716 | 86.9 | 0.716 |
| Allaml | 98.61 | 0.969 | 95.83 | 0.908 | 98.61 | 0.969 | 98.61 | 0.969 | 97.22 | 0.938 |
| Gli_85 | 82.35 | 0.579 | 78.82 | 0.49 | 88.23 | 0.734 | 94.11 | 0.86 | 92.94 | 0.833 |
| Parkingson | 76.45 | 0.387 | 76.58 | 0.397 | 76.45 | 0.387 | 74.2 | 0.36 | 83.2 | 0.53 |
| Prostate_ge | 62.74 | 0.25 | 61.76 | 0.23 | 60.78 | 0.784 | 94.11 | 0.882 | 94.11 | 0.882 |
| Smk_can | 60.42 | 0.211 | 57.21 | 0.148 | 59.89 | 0.2 | 77.54 | 0.548 | 72.72 | 0.453 |
| Yale | 63.03 | 0.603 | 62.42 | 0.597 | 62.42 | 0.597 | 64.24 | 0.616 | 58.78 | 0.558 |
| Gissette | 91.34 | 0.826 | 71.77 | 0.435 | 91.3 | 0.826 | 91.61 | 0.832 | 85.38 | 0.832 |
| Leukemia | 90.27 | 0.779 | 83.33 | 0.632 | 91.66 | 0.809 | 98.61 | 0.969 | 98.61 | 0.969 |
| Colon | 70.96 | 0.402 | 64.51 | 0.255 | 64.51 | 0.255 | 87.09 | 0.723 | 88.7 | 0.74 |
| Madelon | 59.53 | 0.19 | 59.53 | 0.19 | 59.53 | 0.19 | 60.57 | 0.211 | 61.8 | 0.236 |
| Pcmac | 80.03 | 0.601 | 76.58 | 0.531 | 80.13 | 0.603 | 78.17 | 0.561 | 76.89 | 0.535 |
| Base | 90.01 | 0.8 | 83.24 | 0.664 | 90.26 | 0.805 | 83.19 | 0.664 | 81.93 | 0.639 |
| Poker | 50.21 | 0.203 | 50.21 | 0.203 | 50.21 | 0.203 | 50.21 | 0.203 | 50.21 | 0.203 |
| Susy | 73.29 | 0.452 | 73.29 | 0.452 | 73.29 | 0.452 | 72.73 | 0.436 | 74.58 | 0.472 |
| Mobile Health | 45.91 | 0.3 | 45.91 | 0.3 | 45.91 | 0.3 | 54.93 | 0.306 | 54.93 | 0.306 |
| Covid-19 | 79.86 | 0.553 | 79.86 | 0.553 | 79.86 | 0.553 | 80.93 | 0.558 | 80.93 | 0.558 |
| Average | 76.64 | 0.541 | 75.44 | 0.504 | 76.51 | 0.558 | 80.4 | 0.598 | 80.25 | 0.597 |
Table 4 shows the number of features selected by each feature selection algorithm. In addition, the number of comparisons necessary to obtain the subset. In addition, individual results by data set and classifier are placed in tables 5, 6 and 7.
Figure 1 compares the relative accuracy from individual experiments with the test data sets, as given by three different classifiers.
The FSOC algorithm gets less features than CFS, while FCBF and ECMBF select in average more features. Algorithms FCBF y ECMBF are the slowest for large features, followed by CFS and finally by FSOC (see Table 3).
It is interesting to see how FSOC behaves with large data sets (large number of instances), such as Poker, Susy, Mobile Health and Covid-19.
It can be seen that FSOC maintains classification efficiency by reducing the features of large data sets, which tend to generate very large models (mainly tree models) that sometimes cannot be stored in main memory.
The same is true for data sets of thousands of features, where very large and poorly understood models tend to be obtained.
Table 8 Pros and cons for each feature selection method
| Method | Advantages | Disadvantages |
| Correlation Feature Selection (CFS) | Greedy algorithm that obtains an optimal feature set. Maintains and sometimes improves the efficiency of classifiers using selected features compared to using the full set of features. Simple calculations (correlations) that are optimized with matrices. | It requires high processing as it needs to compare multiple features, on datasets of thousands of features it is too slow. Feature search is not exhaustive due to the greedy process it uses. |
| Fast Correlation-Based Filter (FCBF) | Search algorithm is fast when the correlation parameter between features and class is high. A redundancy measure is implemented that is obtained directly from the data and is not manipulated by the user. | A parameter is required by the user to eliminate irrelevant features (α). An erroneously selected parameter would affect the selected features. Based on the statistical approach described in algorithm 3, the number of selected features far exceeds the CFS and FSOC algorithms |
| Efficient feature selection based on correlation measure (ECMBF) | Implements a new measure to relate nominal and numerical features. | It is required to set two parameters; relevance (α) and redundancy (β); assigning these values trivially would imply limited feature selection. Based on the statistical approach described in algorithm 3, the number of selected features far exceeds the CFS and FSOC algorithms. |
| Feature Selection Ordered by Correlation (FSOC) | Fast algorithm for obtaining relevant features. Does not require any assignment of parameters by the user. Simple calculations (correlations) that are optimized with matrices. Ideal for datasets with thousands of features. | Greedy search algorithm, does not perform a global search to obtain the best set of relevant features. In datasets with few features, the speed improvement becomes imperceptible. Correlations calculated could be affected by features with extreme values (noise or outliers). |
Generating models with fewer features helps improve training and validation times, as well as reducing the space required to store models with little or no loss of predictive information.
In addition, if the reduced features are used for tree models, the tree becomes easier to understand. The algorithm with the lowest average in features and computational cost was FSOC.
Although CFS algorithm very slightly exceeds FSOC on the average of percentage of correct classifications and statistical kappa in the three classifiers, it also far exceeds the average of its computational cost compared to FSOC.
The FCBF algorithm has a low efficiency, it is below the rest of the algorithms, while the use of the complete set of features can cause some classifiers to be confused and overfit.
FSOC allows reducing the features to avoid analyzing statistical relationships that are not very discriminatory for the class (target) or the information contained in them are redundant.
In turn, it helps to eliminate those features that are unreliable because they were apparently answered randomly or based on a non-rational critic, helping to reduce data recovery and maintenance costs.
Could further reduction in time be achieved by parallelization? It is possible to further accelerate FSOC by simultaneously computing the correlation between feature
To achieve this with several processors, the complete training set, as well as a fraction of the features, are given to each of them. Each processor will compute the correlation between the features given to it and the class feature
Nevertheless, the rest of the algorithm (lines 10 to 32, algorithm 1) can be run in just one processor, since the time spent by it is short.
5 Conclusions and Future Work
This article presents a method called FSOC that selects relevant features in a way to reduce the computational cost of its selection, with little or no loss of classification accuracy.
Statistical results comparing three well-known methods for feature selection with Feature Selection Ordered by Correlation (FSOC). It measures the computational cost to obtain such reduced set, and the efficiency (number of correct classifications) produced by the selected features.
The efficiency was obtained using classifiers C4.5, Random Forest (decision trees) and Naive Bayes (conditional probability), tested with a collection of 36 data sets available in the open literature.
The results show an efficiency very similar between FSOC and the best algorithm Correlation Feature Selection (CFS), but FSOC is 42 times cheaper with respect to CFS in the computational cost with null or very slight loss of discriminatory power.
Therefore, the FSOC method is especially relevant for high volumes (large data sets) and high dimensionality data (hundreds of thousands of features).
Even though Fast Correlation-Based Filter (FCBF) is fast, it needs to adjust a relevance threshold not to discard useful features. In addition, it classifies with less accuracy, and uses a number of features higher than FSOC does.
The Efficient feature selection based on correlation measure (ECMBF) algorithm is fast, but it is necessary to have a prior knowledge of the data sets, or to find (by trial and error) adequate values for the relevance and redundancy parameters. These extra classifications render it impractical.
Initial work with FSOC on data sets with large amounts of data and high dimensionality (Parkinson, Prostate_ge, Smk_Can, Yale, Gissete, Leukemia, Colon, Madelon, Pcmac, Basehock,Poker, Susy, Mobile Health and Covid-19, Tables 1 and 4, with up to 5,000,000 samples and up to 5,000 features) shows less features selected with no sacrifice in accuracy.
It is planned to perform further testing with additional high dimensionality data sets. It will also be interesting to integrate new ways to discretize numerical features or to find new measures to correlate nominal, numeric and mixed features.











 nueva página del texto (beta)
nueva página del texto (beta)