INTRODUCTION
The ability to engineer microorganisms and thus develop new skills has been a goal for more than half a century, and now it is a reality, thanks to the rise of biological systems in 1990. Nowadays, it is possible to apply the theory of control and cellular program actions such as transcription and translation through genetic circuits (Cameron, Bashor & Collins, 2014). All of this would not have been possible without the work of Francois Jacob and Jacques Monod on the regulation of gene expression through the understanding of the operon lac (Jacob & Monod, 1961), which after a while led scientists to see genetic mechanisms as a circuit that allows cells to respond to their environment. Nevertheless, the concept of genetic circuit is given to the self-regulation of the cell on its genetic material, this through mechanical stimuli such as mechanotransduction. This is a handy tool in tissue engineering because the specific differentiation of a cell towards a particular tissue depends on it (Tadeo, Berbegall, Escudero, Álvaro & Noguera, 2014). For example, the operon lac discovered in 1961 by Jacob and Monod would be considered a natural genetic circuit. However, the natural genetic circuits that control changes in gene expression, morphology, and cell mobility have been selected over time by scientists to improve the fitness of other cells or multicellular organisms outside the original circuit (Meyers, 2015), giving them new characteristics that they did not have before and allowing the rise of synthetic genetic circuits, which will respond to controlled stimuli or signals to induce cells to perform a desired function. Moreover, the response can be monitored by reporters such as the green fluorescent protein (GFP).
Development of a genetic circuit and its components
In order to assemble a genetic circuit, there are specific components that must be taken into account, as described in Figure 1. A genetic circuit can be roughly divided into three parts: 1) a sensor that accepts one or several inputs, 2) internal logic circuit composed so that it can react to the input signal, and 3) an output that can be the protein of interest for industrial/therapeutic applications or act like an actuator for either a second genetic circuit or the molecular mechanism of the cell (Meyers, 2015). The input, which will be the stimulus that activates the “biosensor”, can be mechanical, chemical, physical (light, heat), antigens, nucleic acids, or the use of other proteins in an internal self-regulating system (inside the cell). The internal logic circuit or processor, which will be the necessary components for the operation of the latter, is composed of the basic structure of a gene: a promoter, a ribosome binding site (RBS), an open reading frame (ORF) or coding sequence (in synthetic circuits, this will facilitate the expression of the desired protein in case the chassis organism does not have the necessary mechanisms to make the functional protein), and a terminator (in order to stop the mechanism of the genetic circuit). The outputs of the circuit are frequently reporters like fluorescent proteins that can be easily detected and quantified, or it could be an actuator for a second circuit if the output of the first circuit is a small molecule that can induce a response and act like an input for the second biosensor.
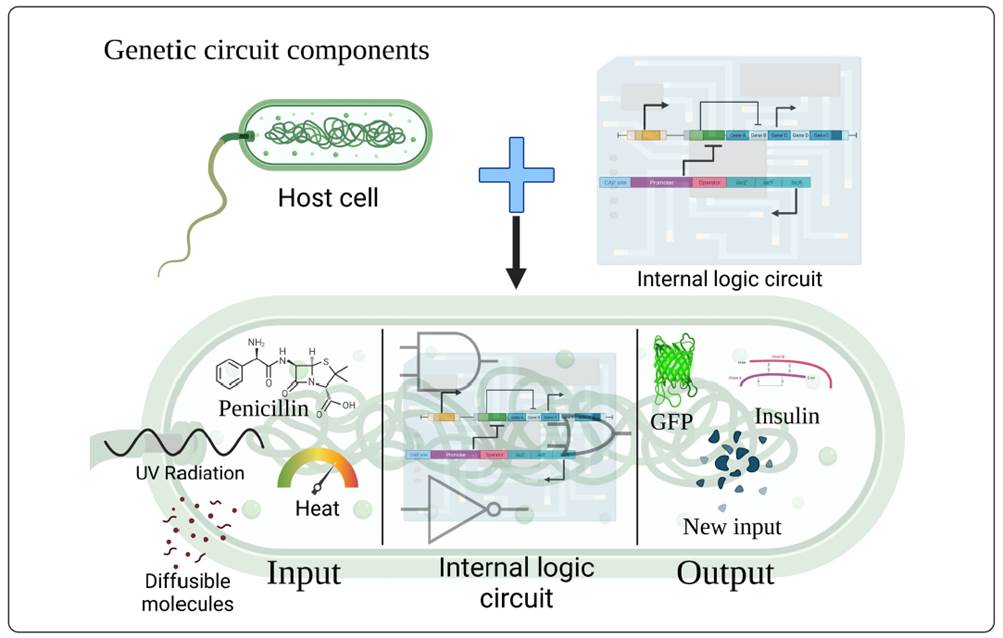
Figure 1 General composition of a synthetic genetic circuit, where the input is found at the left side of the cell. Inputs can be antibiotics (Penicillin), diffusible molecules that the bacterium uses to communicate with others, and even external stress such as heat or UV radiation. The internal logic circuit will be where the interactions between the stimulus and the genes will take a Boolean behavior to give rise to an output. The output may be a protein of interest (Green Fluorescent Protein GFP or Insulin for instance) or a new input from another external circuit or this same circuit in order to generate feedback, either positive or negative. Modified of (Cui et al., 2021; Kitada, Diandreth, Teague & Weiss, 2018; Wang & Buck, 2012).
All these components (input, processor, and output) can be put inside a host cell forming a synthetic organism and, in the future, has industrial applications such as the synthesis of therapeutic proteins, improving the yields in the production of biofuels, and controlling cell mechanisms (proliferation, differentiation, and morphogenesis, for instance) (Meyers, 2015). These applications will be described in the next sections.
Chassis or host cell
In most research, model organisms are used as a biological chassis because their genome is well known; for that reason, in research with bacteria, the organism Escherichia coli is used. For example, in 1982, the effect that UV radiation could have as a switch for a natural genetic circuit was already being investigated, the experiment consisted in the induction of the lytic route of lambda phage through a 45-minute exposure after infection (Ptashne, Johnson & Pabo, 1982). On the other hand, for genetic circuits in eukaryotes, the Saccharomyces cerevisiae yeast is used. For example, it was applied in enhancing biofuel production such as n-butanol by modification in the actin cytoskeleton in order to increase both the cell growth and production of n-butanol and medium fatty acids, resulting in the highest n-butanol titer of 1,674.3 mg L-1 ( Liu et al., 2022; Meyers, 2015). Nonetheless, there may be problems with the use of foreign host cells, as the genetic circuit and the host cell cannot be 100% compatible most of the time. For example, the synthetic biologist Chris Voight made a genetic circuit with parts of the bacterium Bacillus subtilis. This would have applications for the regulation of the activity of specific genes against chemical stimuli as an input signal (Kwok, 2010), but he wanted to study this circuit independently from the original microorganism, so he inserted the circuit into E. coli. Unfortunately, despite the genetic circuit being well designed, it failed and the new chassis collapsed because the circuit was incompatible with part of the biological machinery of the host cell. However, not all was lost because a new technique called “Codon Optimization” had been developed. This technique allows the structure of the foreign gene to be reconfigured so that it is compatible with the new chassis (Elena, Ravasi, Castelli, Peirú & Menzella, 2014). This reconfiguration requires the search for homologous sequences to the most frequently used foreign codons in the new chassis (E. coli) using bioinformatics. Then, the optimized sequence is obtained, synthesized, and inserted into the vector or our circuit using DNA Assembly techniques such as Gibson Assembly (Gibson et al., 2009) or BioBrick Assembly (Polizzi, 2013) for subsequent use in the production of drugs, molecules of interest, or fuels.
Inputs of synthetic circuits
Many genetic circuits are designed to respond to a specific stimulus; these can be antibiotics, diffusible molecules that the bacteria use to communicate with each other, and even external stress such as heat or UV radiation (Meyers, 2015). The use of the latter is recommended because the damage is less invasive and presents good reversibility compared to the cytotoxicity that can be caused by other inputs such as chemicals or metals (Liu et al., 2018).
In order not to cause disturbances within the cell, the control elements or inputs to the genetic circuit must be foreign or non-existent within the cell, and the correct use of a series of inputs to form Boolean logic gates can help dictate the timing, duration, and functionality of the circuit (MacDonald & Deans, 2016).
Outputs of synthetic circuits
The output of the circuit would be the protein of interest or one that serves as an input to another external circuit or this same one in order to generate feedback, either positive or negative. Positive feedback can have a “snowball” effect. This is because it can intensify small signals, while the negative feedback is responsible for reducing noise within the system and making the response or output of this system more stable (Kassaw, Donayre-Torres, Antunes, Morey & Medford, 2018). In genetic circuits, most of the outputs present a fluorescent or reporter molecule. After all, this makes its quantification more straightforward by techniques such as flow cytometry (Meyers, 2015). The GFP protein can be removed from the protein of interest to prevent it from negatively influencing its activity by the enzyme enteropeptidase (Martemyanov, Shirokov, Kurnasov, Gudkov & Spirin, 2001). It is expected that in the future, the possible outputs of a genetic circuit will be of great interest from an industrial point of view. Proteins that produce changes in cell morphology, specific cell differentiation (tissue engineering), or synthesis and secretion of therapeutic proteins will be further developed in the following topics.
Genetic integrated circuits and genetic memory
An integrated circuit in electronics is composed of several logic gates that interact with each other to fulfill specific functions; mimicking this term for genetic circuits, these would be several independent circuits acting as logic gates, and the input signal would promote their responses. The behavior that the parts of the circuit adopts according to the input(s) will be what classifies it as a specific logic gate (AND, OR, NAND, NOR, etc.), and these behaviors can be permanently recorded in the DNA of the microorganisms through the use of recombinases. Genetic biologists have used these recombinases for years in order to generate knockout and transgenic mice, and now, synthetic biologists have found them useful for genetic circuit production (MacDonald & Deans, 2016). During the last years it has been achieved that microorganisms that possess this type of circuit can develop a degree of “genetic memory” (Figure 2), as well as that the functionality of the circuit is transmitted from generation-to-generation (Nandagopal & Elowitz, 2011) and in a stable way. According to works such as Becskel’s in Germany (Becskel & Serrano, 2000), how the negative feedback of an artificial genetic circuit increased its stability in E. coli compared to an unregulated circuit was demonstrated. However, the stability of feedback loops can be compromised if the repression factor is low and their life is not very high. Also, the type of feedback can have a strong correlation with the genetic loop; genetic loops are based on biochemical interactions within the confined space of the cell (Moon, Lou, Tamsir, Stanton & Voigt, 2012), so, interaction with the cell is impossible to avoid or rather to ignore, but these interactions can be beneficial. For example, while the negative feedback offers us the stability of the circuit, positive feedback can help us manage specific reactions at the cellular level such as modulating the growth of our host cell, offering an emergent bio-stability due to these interactions (circuit-host), and at the same time offering a stable switch. These results were in accordance with the works by Tan, Marguet & You, (2009) on E. coli with this type of feedback on a circuit that modulates the production of T7 RNAP, which is a mutant RNA polymerase that regulated cell growth and offered these bio-stability properties.
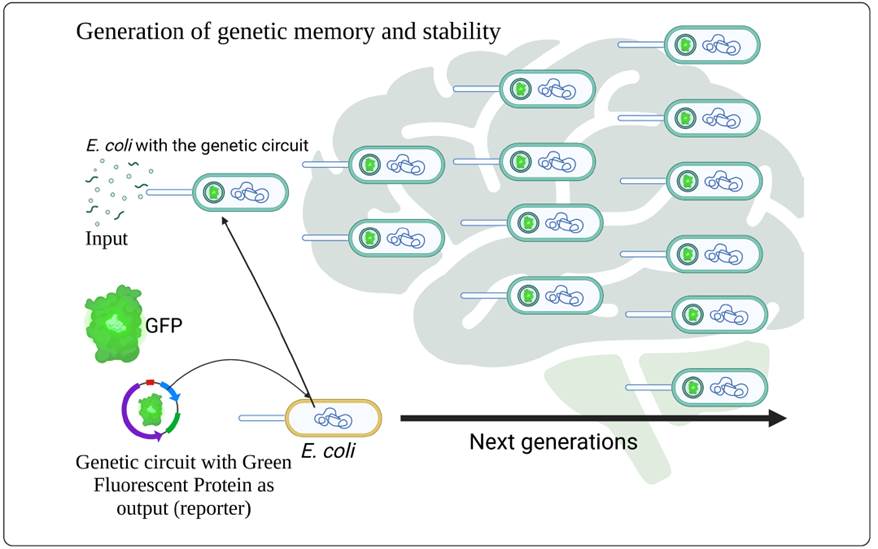
Figure by personal creativity.
Figure 2 (Genetic memory): Stability of the memory generated by a genetic circuit introduced into E. coli (yellow cell) that responds to an input and through the interactions that this generates within the internal circuit (Plasmid with Green Fluorescent Protein) is able to generate a series of actions that will allow the next generations (green cells with GFP plasmid) to “remember” these interactions and maintain the main function of the circuit that in this case is the production of GFP for several generations without the need to be exposed to the input or stimulus again.
As far as genetic memory is concerned, this refers to the use of a specific metabolic pathway after a particular time. Genetic memory occurs thanks to the recognition of signals or inputs that interacted with our chassis in the past, allowing the use of the genetic circuit and preventing it from being lost after a few activations. For example, in the use of bioreactors, transitory conditions such as growth phase or substrate concentration can be converted into permanent signals that act as a stimulus to turn on the genetic circuit, so that each time these conditions are repeated over time, the circuit can “remember” these stimuli and activate itself, maintaining its effectiveness over time.
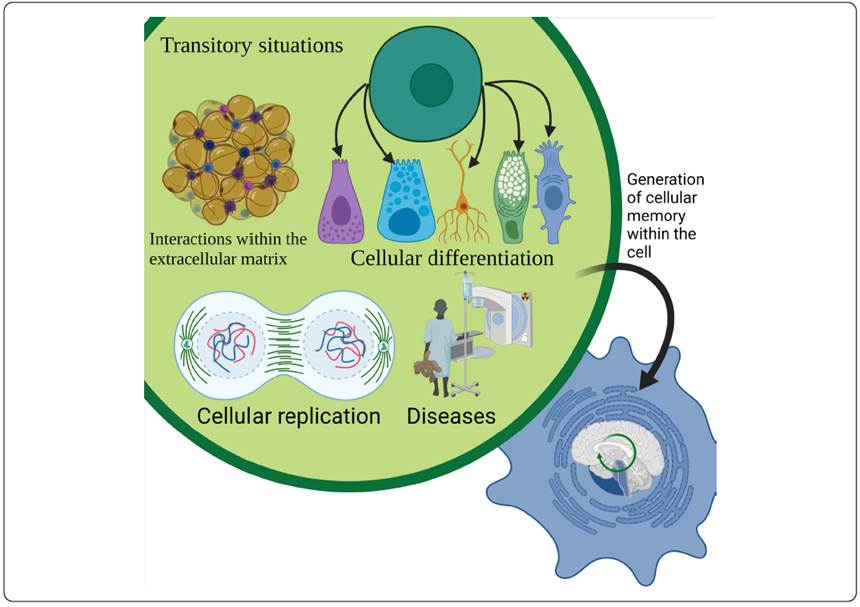
Memory can even be generated for more transcendental conditions that are difficult to repeat or remember under in situ conditions (Yang et al., 2014). This memory can be achieved through the cell’s ability to convert transient signals such as changes in extracellular matrix (ECM), presence or absence of other cells, and protein levels as they experience differentiation, healing, and disease progression into a memory encoded in the genome (Figure 3), allowing synthetic biologists to track various biological phenomena at the molecular level (MacDonald & Deans, 2016). This cell memory can also be generated by feedback in the circuit, allowing for stability, even if the feedback or input is no longer present. With the use of logic gates, a circuit can also be developed that remains active even if the input is not present, allowing its stability and memory for up to 90 generations. That memory and stability were achieved through the use of recombinases, which surrounded parts of the circuit that allowed the behavior of a specific logic gate, depending on which part was altered by the recombinase, through the inversion of promoters or ORF (Siuti, Yazbek & Lu, 2013).
Modeling of genetic circuits
To design and implement an effective control system, it is necessary to describe the behavior of the process to be controlled. The representation of this behavior is named as a modeling system. In general, the dynamics of the model is represented in mathematical terms, but it may be represented in many different ways depending on the control objective and the particular process (Ogata et al., 2010). Modeling is essential for both the analysis of natural systems and the design of synthetic ones. For instance, it can help predict the behavior of a genetic circuit at the production level in synthetic biology. However, several problems arise in modeling cellular circuits: parameter sensitivity, simplification of complex circuits, and extrinsic noise (Sprinzak & Elowitz, 2005). In engineering, system modeling helps predict how a system will behave, providing a close approximation to reality and supporting the design, control, and optimization of processes (Dunn, Heinzle, Ingham & Prenosil, 2013). In synthetic biology, mathematical modeling can help predict how a genetic circuit will behave at the production level.
Why the use of mathematical models?
The dynamics of many systems are described in terms of differential equations using physical laws governing particular systems. Nevertheless, the mathematical model obtained must have a compromise between the simplicity of the model and the accuracy of the results. It means that modeling is a simplification and does not include all aspects of the problem and is generally only valid in certain situations. However, the following advantages of mathematical modeling can be summarized as follows:
Modeling applications in synthetic biology and biosciences
The use of modeling for mathematical processes in biosciences can be visualized from the enzymatic reaction to bioreactor control, for example, modeling the production of Docosahexaenoic Acid (DHA) (Song, Zhang, Kuang, Zhu & Zhao, 2010), antibiotics such as pleuromutilin (Benkortbi, Hanini & Bentahar (2007), or gluconic acid fermentation in batch-type reactors (Liu, Weng, Zhang, Xu & Ji, 2003). Furthermore, the Gompertz model can predict the cell growth rate and lag phase (Ardestani, Rezvani & Najafpour, 2017; Buchanan & Cygnarowicz, 1990; McKellar, 1997; McKellar & Knight, 2000; Mechmeche et al., 2017; Perni, 2013; Tjørve & Tjørve, 2017). According to the above, the cell is taken as a factory that can produce chemical compounds, biogas, or in the case of synthetic biology, the product of interest such as insulin (Zhang & Tzanakakis, 2019), specific antibodies (Hynes & Lane, 2005), or messenger RNAs with the 4 cell reprogramming factors (Takahashi et al., 2007). The use of accurate modeling helps represent the genetic circuit in such a way that a gene regulation system can be designed to produce proteins whose concentrations always oscillate due to their degradation in a given time or to design a circuit that produces as much ethanol as possible with a certain amount of substrate, glucose, and so on (Baldwin et al., 2012).
Another application of modeling in biosciences is to predict cancer development (Jonkers & Derksen, 2007). In 1984, a mathematical model for the growth of breast cancer was proposed (Botesteanu, Lipkowitz, Lee & Levy, 2016), and it was found that different individual tumors grew according to Gompertz parameters. With this basic growth model, it was possible to estimate that a cancerous cell would take eight years to be detected, referring to the fact that a tumor is a group of cells that grow without control and its growth is limited to the space and resources available (Patel & Nagl, 2006), which makes it more complicated to describe its growth and behavior as opposed to growth under an exponential model. Hence, from that day forward, the use of the Gompertz model has been widely used in cancer research.
Applications in the biomedical and energy industry
Once the design and operation of the circuits, in general, are well understood, what follows is to know how to assemble them in such a way that they fulfill a function of our interest. For example, mimicking the electronic circuits, if we take a transistor that has a specific function, the transistor is not limited to perform a different function or behavior in conjunction with other parts (other transistors, logic gates, capacitors and so on) that by itself could not. By the same way, as more complex electronic circuits can be deve loped, we can develop more complex genetic circuits with new functions. Three stages have been proposed to construct a synthetic biological system (Muñoz-Miranda et al., 2019):
Decoding and analysis of the genomes of different biological systems.
Synthesis of the parts that make up a genome and its subsequent assembly.
Use of genome editing tools.
In this way, genetic circuits with exciting and unlimited future applications can be designed. An example was proposed by Jay D. Keasling, Ph.D., who has had one of the most recognized achievements in this area of knowledge. This example was the synthesis of the artemisinin precursor and artemisinic acid by improving the mevalonate pathway using a synthetic circuit inserted in S. cerevisiae in order to create this drug for the treatment of malaria (Ro et al., 2006). This is one of the many applications that can be used to exemplify the use of genetic circuits in the biomedical field, as well as the synthesis of antibodies through synthetic biology (Paduch & Kossiakoff, 2017). Following the three steps described above, scientists can take the desired antibody sequence and synthesize it together with a genetic circuit in order to insert it into a chassis for its expression. This procedure seeks to increase yields compared to the hybridoma technology developed by Köhler & Milstein (1975) or terpenoid synthesis, likewise as a treatment for malaria using the mevalonate pathway with S. cerevisiae as a chassis or using them as an anti-cancer drug (Martin, Piteral, Withers, Newman & Keasling, 2003).
Synthetic biology in microorganism for the treatment of diseases
Since ancient times, biotechnology has allowed microorganisms or parts of them to obtain products of high interest and benefit society. It began with fermentations to obtain wines and bread and has developed in such a way that they are now used for the production of more environmentally friendly biofuels or the treatment of diseases directly in human cells with CRISPR-Cas9, such as Leber congenital amaurosis 10 (LCA10), which causes blindness, by eliminating the mutation in the CEP290 gene that is responsible for LCA10. The use of CRISPR-Cas9 has a more significant advantage than virus therapy because this technique is more versatile and easy to design. The CEP290 gene is too long to introduce into a viral genome (Ledford, 2020).
Synthetic microorganisms have been developed for the treatment of diseases such as diabetes, with cells being obtained that produce insulin by exposure to blue light that were tested on diabetic mice to demonstrate improved health status (Zhang & Tzanakakis, 2019), or for the treatment of cancer, such as the use of Salmonella typhimurium, because some bacteria are known to have the capacity to agglomerate in tumors. Synthetic S. typhimurium expresses the GFP reporter to monitor how the bacteria infect xenografts, inhibit tumor development, and cause tumor regression ( Zhao et al., 2005). In another study, a lab worked with a non-infectious Salmonella sp. to express anti-cancer proteins using a high-density dependent expression system that uses quorum-sensing. When the Salmonella population increased in the tumor (which is a closed space), it emitted a switch-like signal that activated a positive feedback loop or snowball (Swofford, Van Dessel & Forbes, 2015), leading to the inhibition of ErbB receptors, which have aberrant expression and overexpression in various types of cancer (Ho, Moyes, Tavassoli & Naglik, 2017). However, by using specific immunoconjugate antibodies such as 528 IgG-rRA, it is possible to have a cytotoxic effect on A431 cells by binding to ErbB receptors, causing tumor cells to slow down their proliferation and protein synthesis and then die (Hynes & Lane, 2005; Masui, Kamrath, Mendelsohn, Apell & Houston, 1989; Sato et al., 1983). The application of synthetic biology makes it possible to optimize the production of various products, facilitate downstream processes in bioprocesses, improve chassis with better tolerance to stress situations, and allow excellent stability of these circuits to express complex pathways. E. coli is used to express approximately 30 genes stably, including insulin and vitamin B12. Within the biomedical applications, there is the development of an E. coli strain capable of invading tumor cells and releasing interfering RNAs so that the expression of the CTNNB1 gene is knocked down. The over-expression of the oncogenic mutation of this gene is involved in the development of several types of colon cancer, an example of which is done in the engineering of the microbiome (Ruder, Lu & Collins, 2011). There are also applications in regenerative medicine using synthetic RNAs that translate four cell reprogramming factors: Oct3/4, Sox2, Klf4, and c-Myc, described by Takahashi and Yamanaka (Takahashi et al., 2007), without the need to express other genes, for example, with the use of viruses. This is important because the additional copies of the four genes, permanently inserted into a genome, can make the cells more prone to mutations or even the generation of cancer. Thus, synthetic RNAs are faster and have better performance of the process (Ruder et al., 2011).
The development of synthetic biomolecules can be significantly exploited, such as the creation of artificial enzymes by protein hybridization (Villarino et al., 2018) to treat the recent outbreak of coronavirus 2019-nCoV (COVID-19), a highly contagious disease related to a market in Wuhan, China where several people presented pneumonia of unknown causes and that has already infected at least 1,975 cases by January 25, 2020 (Wu et al., 2020). The virus could be obtained for characterization from the bronchoalveolar fluid of positive patients, followed by RNA extraction and sequencing (Zhu et al., 2020). The 2019-nCoV sequences are 96% identical to those of the coronavirus in bats and 79.6% similar to those of SARS-CoV, which has a mortality rate of 9.6% and has infected 8,096 people and caused 774 deaths (Wong et al., 2015). SARS-CoV has a mortality rate higher than 2019-nCoV; it was 3% at the beginning of the year, although by March and April, it was already 3.61% (Khafaie & Rahim, 2020) and 6.30% worldwide (Bulut & Kato, 2020), respectively. This is because unlike SARS-CoV, this new coronavirus (2019-nCoV) can grow better in primary human respiratory cells than in normal tissue culture cells (Perlman,2020), and furthermore, analysis of the structure of the spike protein (S) of 2019-nCoV shows that it binds weakly to the human ACE2 receptor, while the SARS-CoV S protein did have a strong affinity (Dong et al., 2020). SARS-CoV-2 utilizes a highly glycosylated homotrimeric spike protein (S) (consists of S1 and S2 subunits) for receptor binding and virus entry. Besides, to engage host cell receptor human angiotensin converting enzyme 2 (hACE2), S protein undergoes dramatic conformational changes (Lan et al., 2020). The diagnosis of 2019-nCoV can be carried out by real-time RT-PCR (Reverse Transcription Polymerase Chain Reaction) (Corman et al., 2020; Holshue et al., 2020), which is more sensitive than conventional RT-PCR, thus having a precise diagnosis in early infections (Shen et al., 2020). Also, for multiple samples, ELISA (Enzyme-Linked Immunosorbent Assay.) or GICA (Colloidal Gold Immunochromatographic Assay) can be used (Xiang et al., 2020). The role of synthetic biology includes the treatment of 2019-nCoV using virus sequences with which it is possible to synthesize antibodies that block the binding between SARS-CoV-2 and the human angiotensin II receptor (hACE2). This is recent work, and it promises to be an effective treatment while a vaccine is developed. Samples were taken from patients who expired the COVID-19 in search of specific Ab that could block the binding of SARS-CoV-2 with hACE2 by ELISA assays to determine this blockage. Three samples achieved a strong blockade of SARS-CoV-2 RBS towards hACE2; from here, the variable regions of IgG’s heavy and light chains were amplified by reverse PCR for subsequent cloning. In the blockade test of SARSCoV-2 RBS towards hACE2, two of these monoclonal antibodies called 311mab-31B5 and 311mab-32D4 exhibited high blocking efficiency in both ELISA and the confirmation by flow cytometry (Chen et al., 2020). This is the first work of this type but not the only one expected. For example, Jacoc Glanville, Ph.D. works with the SARS-CoV sequences using DNA shuffling, and he is looking for a variant that achieves blocking effects on 2019-nCoV. There is also the design of synthetic peptides or aptamers (Haußner, Lach & Eichler, 2017) that can induce an immune response mediated by CD4+ and CD8+ cells through an epitope-parotope interaction (Bojin, Gavriliuc, Margineanu & Paunescu, 2020); or nano-vaccines based on nanoparticles such as VLPs (virus-like particles) acting as delivery vehicles of the antigens that trigger protective immunity by inducing the generation of antibodies to fight the virus (Palestino, García-Silva, González-Ortega & Rosales-Mendoza, 2020); or a plasmid with synthetic DNA of the coronavirus S protein, as was done with MERS-CoV (Muthumani et al., 2015); or the application of new diagnostic techniques such as microfluidics, which were used for the SARS-CoV virus (Kim, Marafie, Jia, Zoval & Madou, 2006; Zhou et al., 2004). All these treatments promise to be more effective than the use of angiotensin 1 receptor blockers (AT1R) and ACE. It is hypothesized that the administration of these blockers may increase the risk of developing a severe case of COVID-19 (James, 2020), as those blockers can increase the number of hACE2, which is the entrance of the virus into the body. Nonetheless, that may be beneficial, as the cellular machinery that the virus needs to reproduce would be busier producing hACE2 than copies of the virus (Gurwitz, 2020). Both theories require a greater understanding of the mechanism of these blockers in the human body. However, only one blocker (olmesartan) has shown an increase in hACE2. Other blockers such as losartan, candesartan, valsartan, and telmisartan have no increase in this receptor (Speth, 2020). Their use has been proposed because AT1R produces inflammation and its blockers (ARBs) can decrease inflammation and reduce the respiratory complications caused by the virus, although so far, there is not enough evidence to say that these blockers have a direct effect on the complications or the output of COVID-19 (South, Tomlinson, Edmonston, Hiremath & Sparks, 2020; Thomson, 2020). In experimental models of acute lung injury, including a model of SARS-CoV infection, scientists have observed that angiotensin-receptor blocker (ARB) may mitigate COVID-19 by attenuating Ang II-mediated acute lung injury by blocking the type 1 angiotensin receptor (AT1R) (Kuba et al., 2005). More direct use of well-developed synthetic biology and not only the use of some tools like molecular techniques would be a deep dive of these interactions between the virus and human receptors as an input for some therapeutic process in the form of a genetic system or circuit, but time will allow us to unveil more significant applications of this science in future problems and to be ready for them.
Synthetic biology in microbiome engineering
The microbiome is the community of microorganisms and genomes associated with the meta-genome of the human body (Galloway-Peña, Brumlow & Shelburne, 2017). It has an essential role in the organism; for example, it can regulate the immune system or have effects against pathogens (Buffie & Pamer, 2013). Direct control of the microbiota can have beneficial effects even from an infection against cancer (Galloway-Peña et al., 2017), where organisms such as E. coli and Pseudomona aeruginosa, for instance, may predominate (Gedik et al., 2014) and lead several complications such as pneumonia in the case of a predominant infection of P. aeurignosa (Evans & Ost, 2015). The microbiome also affects the immune system through small molecules such as short-chain fatty acids as Butyrate that suppresses NF-κB signaling through downregulation of TNF-α or Clostridium sp. that may induce the secretion of anti-inflammatory cytokines such as IL-10 (Dou & Bennett, 2018).
In these interactions between the microbiome and the human body, it is not surprising that disturbances in the composition of the microbiome could lead to complications in its correct functioning or even develop problems such as colon cancer through peptides released by the microbiota such as Phr0662 from Bacillus sp. or EntF-metabolite from Enterococcus faecium (Arnold, Roach & Azcarate-Peril, 2016; Kang & Martin, 2017). Colon cancer can be diagnosed by the microbiome itself because these interactions are so direct that a metagenomic analysis of the fecal microbiota can be used as a diagnosis of this disease (Zeller et al., 2014). For cancer treatment, some microorganisms of the microbiota are used, such as Bacillus polyfementicus that can stimulate the inhibition of ErbB2 and ErbB3 (Sun, Gajurel, Buys & Yin, 2019). Microbial communities have better stability for plasmid prevalence and exogenous DNA spread (Sheth, Cabral, Chen & Wang, 2016), making them good chassis for hosting genetic circuits that can fight diseases (Ruder et al., 2011). Some of the symptoms of chronic colitis are known to be caused by tumor necrosis factor alpha (TNF-α), and one therapy is the intravenous application of the anti-TNF-α antibody. However, this treatment is expensive and triggers side effects, so nanobodies, which are derived from the heavy chain of antibodies from dromedaries or camelids, are being developed, and these molecules can be easily cloned into bacteria and yeast or microbiome for the treatment of this disease (Vandenbroucke et al., 2010).
Synthetic biology is also being applied against cholera, a disease caused by Vibrio cholerae and characterized mainly by the passage of bulky stools that lead rapidly to dehydration (Kaper, Morris & Levine, 1995). V. cholerae secretes a small number of cholera toxins (CT) during infection because the same bacterium has self-inducing signals to count the number of toxins released. Thus, when the levels of CAI-1 and CAI-2 (self- inducing cholera 1 and 2) are high, the expression of virulence factors stops. The understanding of that “switch” on the expression of virulence factors helped to modify an E. coli bacterium that could produce CAI-1. When an infant mouse ingested the modified E. coli 8 h before infection with V. cholerae, the survival rate increased dramatically and the intestinal attachment of the toxin was reduced by 80% (Figure 4) (Duan & March, 2010; Ruder et al., 2011). In order for the synthetic E. coli chassis to be well assimilated by the host organism (the infant mouse), the E. coli Nissle 1917 strain, which was isolated from an officer of the Second World War who did not suffer from any intestinal disorder, was used (Schultz, 2008). Since then, this strain is used as a probiotic and is a right chassis candidate for human use, with applications for immunotherapy. In this strain of E. coli, a genetic circuit has been developed to synthesize nanobodies for PDL-1 and CTLA-4 (which are regulators of the immune system that cancer cells use to prevent it from acting on them (Buchbinder & Desai, 2016)) and subsequently release them into cancer cells by means of quorum-sensing, which causes the circuit to induce the lysis of the bacteria and release the nanobodies (Gurbatri et al., 2020). This has been tested on mice and showed promising results.
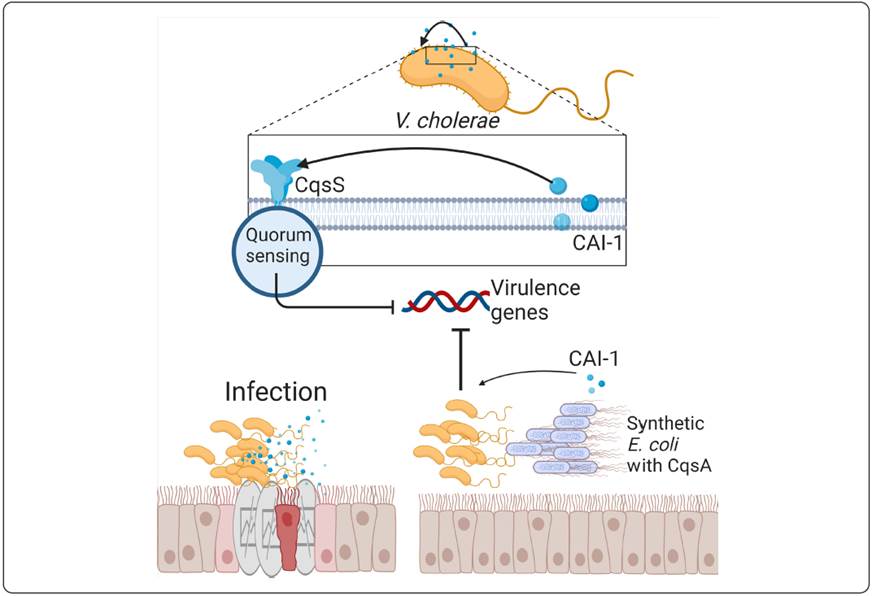
Figure by personal creativity.
Figure 4 Generation of a genetic circuit by quorum sensing for cholera treatment in mice. By using several synthetic E. coli that expressed CAI-1, it was possible to use the regulation machinery of V. cholerae and thus achieve the reduction of the disease produced by this pathogen in mice that were used for this experiment.
Synthetic biology and biofuels
Society’s strong dependence on fossil fuels as the primary energy source has had a significant adverse effect on the environment and human health (Patel, Lee & Kalia, 2018). The depletion of fossil fuel reserves, unstable prices, and increased pollution have led to a search for new energy sources (De Koning, Huppes, Deetman & Tukker, 2016). Within the new renewable energy sources, there is the use of biomass with some nuclear energy in order to compensate for the energy demand that could not be fully covered by the current renewable energy technologies such as biomethane, biodiesel, and biohydrogen (De Koning et al., 2016). Biofuels would limit the average global temperature increase to less than 2 °C, causing carbon dioxide emissions to decrease by 70% by 2,060 compared to 2,014 levels (Oh, Hwang, Kim, Kim & Lee, 2018). An approximation of this reduction of greenhouse gases is based on the effects of 2019-nCoV in China. Only in the first two months of the year, carbon and NO2 emissions have been reduced by 25% and 30%, respectively, by social isolation and reduction in car traffic (Tahir & Masood, 2020). Biohydrogen is standing out among the other biofuels because it has zero contaminants because its only waste during oxidation is water (Oceguera-Contreras et al., 2019). Besides, it is the one with the highest energy content (120 kJ/g), approximately three times more than the energy content from fossil fuels such as gasoline with 41.2 kJ/g or diesel with 42.9 kJ/g for instance (Sekoai et al., 2019). The yield of hydrogen production will depend on the type of microorganisms used and the final products of fermentation. Only one-third of the substrate can be used for hydrogen production; this is because two thirds of the substrate will form small acids, e.g., acetic and isobutyric acid (Zhao et al., 2013), when this process is carried out in a bioreactor, the yield of hydrogen production will now also depend on specific operating conditions such as the concentration of the substrate, pH, temperature and hydraulic retention, in order to obtain the highest possible yield (Oceguera-Contreras et al., 2019; W. Zhao et al., 2013). The most used method for the bioproduction of hydrogen is dark fermentation; this type of process is very environment friendly because it can be carried out with organisms and substrates that are easy to find. This process apparently can be optimized with nanotechnology and synthetic biology. The presence of nanoparticles, for example, the use of Au NPs with a size of 5 nm, could optimize the dark fermentation by 56% (Zhang & Shen (2007), while Ag NPs showed promising results in both lag phase reduction and hydrogen production at a concentration of 0.7 gN/L, obtaining a yield of 190.7 mL of hydrogen. The yield increase seemed to be directly proportional to the concentration of NPs. However, at 1.1 gN/L concentration, the yield decreased to 112.9 mL, showing toxicity in the microorganisms (Zhao et al., 2013). Iron nanoparticles at 400 mg/L were used and compared with a control group to observe if there were changes in the microbial composition, and it was found that the control group contained mostly microorganisms of the genus Enterobacter and small groups of the genus Clostridium, while the consortium treated with the zero-valent Fe NPs were dominated by microorganisms of the genus Clostridium, which have higher yields in hydrogen production (Patel et al., 2018) (Figure 5).
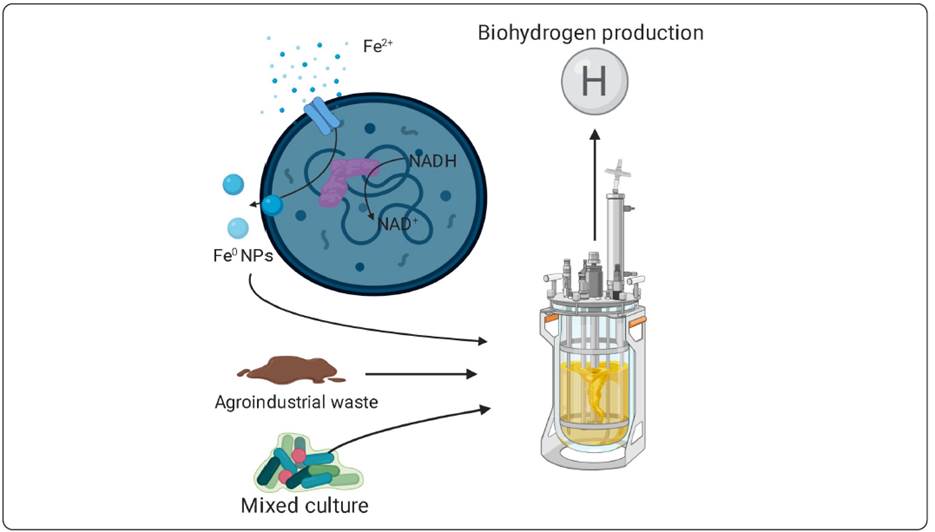
Figure by personal creativity.
Figure 5 Hydrogen production can be optimized by new technologies such as nanotechnology and synthetic biology acting directly on hydrogenases, such as Fe NPs, which can increase electron transfer from hydrogenases such as Clostridium acetobutylicum . This has been shown to work well in E. coli by implementing it in a synthetic route.
Another way to optimize the function of hydrogenases is through genetic modification of hydrogens. Hydrogenases that re-oxidize the hydrogen produced present a problem when trying to obtain fair values of accumulated hydrogen. Therefore, in several studies, an increase in hydrogen production was observed when mutating these hydrogenases or directly eliminating this type of hydrogenase or, on the contrary, inserting another type of hydrogenase, which can improve the capacity of hydrogen production, for example, when expressing the hydrogenase [Fe-Fe] of the anaerobically strict bacterium Clostridium pasteurianum within the cyanobacterium Synechococcus sp. This experiment, besides demonstrating the capacity of this cyanobacterium to accept foreign hydrogenases, resulted in a significant increase in the production of hydrogen, as well as to accept more oxygen tolerant hydrogenases such as Ni-Fe (Patel et al., 2018). The Agapakis group took a hydrogenase [Fe-Fe], a ferredoxin and the pyruvate ferredoxin oxidoreductase (PFOR) for the production of hydrogen through the breakdown of glucose into E. coli as a chassis. This procedure required genes from several hydrogenases such as the ones from Chlamydomonas reinhardtii and the ferredoxin I gene from Spinacia oleracea. These genes went through Codon Optimization for expression in S. cerevisiae and thus were acceptable for use in E. coli. The hydrogenase genes of Clostridium acetobutylicum and Clostridium saccharobutylicum were cloned from plasmids, and the hydrogen genes HydA and HydB were cloned by PCR from Shewanella oneidensis colonies. Other ferredoxin genes were taken similarly from C. acetobutylicum, and the PFOR gene was isolated from the plasmid pLP1 of Desulfovibrio africanus. The fusion of all these genes for hydrogenase and ferredoxin proteins was achieved by the BioBrick standard assembly technique, and the multiple cloning sites were mutated to accept more BioBrick. In this experiment, the highest hydrogen production was seen with C. acetobutylicum hydrogenases and ferredoxins and PFOR of D. africanus. This experiment demonstrated how these synthetic systems work not only to optimize hydrogen production but also to better understand the electron transfer system, which opens the way to the development of better systems through synthetic biology and thus the design of better strains of E. coli that can express synthetic pathways of higher industrial interest (Agapakis et al., 2010).
Other projects in the energy field have been developed with the use of synthetic biology, like the synthesis of n-butanol within S. cerevisiae with an armed route of isoenzymes from different microorganisms (S. cerevisiae, E. coli, Clostridium beijerinckii, and Ralstonia eutropha), which were replaced by Clostridial enzymes (Steen et al., 2008). Also, second-generation biodiesel can be produced from biomolecules such as hemicellulose (Steen et al., 2010).
Thanks to the synthetic biology that allows the optimization of the metabolic routes and the host cell, achieving new routes or the synthesis of nanomaterials such as metallic nanoparticles for the optimization of hydrogen production, the use of fossil fuels can be discarded shortly (Patel et al., 2018; Peralta-Yahya, Zhang, Cardayre & Keasling, 2012).
Conclusions
The application of synthetic biology in the biosciences as well as in the energy industry has a promising future thanks to the types of circuits and techniques that are being developed. In the future, the applications of synthetic biology could be unlimited, from agriculture, regenerative medicine, microbiome engineering, and new generation biosensors to new challenges such as high greenhouse gas emissions or new pathogens such as the SARS-CoV-2 virus that now keeps us on health alert. Also, important will be the development of new techniques that evolve in such a way that their prices are more accessible, such as sequencing technologies and DNA Assembly techniques that are faster, more effective, and easier to use.











 nueva página del texto (beta)
nueva página del texto (beta)