Introduction
Production of bananas and plantains (Musa sp.) is a significant income in countries with tropical climate in Latin America, the Caribbean, Africa and southeastern Asia. In 2018, global gross export is estimated in 19.2 millions of tons (FAO 2018). In 2018, Mexico produced 2.8 millions tons in Chiapas, Tabasco, Veracruz and other states of the Pacific coastline such as Oaxaca (SIAP 2018). Out of the 2.8 millions of tons produced in Mexico, 0.5 were exported, but exports increase year after year because of a trade agreement with China (Gobierno de México 2020).
The Panama disease, caused by the ascomycete fungus Fusarium oxysporum f.sp. cubense (Foc) is the more influential factor that has affected the banana production (Ordonez et al., 2015). Based on the banana host cultivar infected, Foc is classified into 4 races, being Foc tropical race 4 (FocTR4) (aka Fusarium odoratissimum) the most aggressive one that infects a broad range of cultivars, including plants from the Cavendish group, the most consumed cultivar world-wide (Fourie et al., 2011). FocTR4 was recently identified in Colombia (Garcia-Bastidas et al., 2020). The fields of Mexico have been free of FocTR4, but the dispersion of FocTR4 in Latin America is unavoidable, which implies future decrease in banana production.
Trichoderma comprises soil-borne and endophytic fungal species with antibiotic and mycoparasitic activities (Mukherjee et al., 2013) Trichoderma spp. also facilitate nutrient uptake, produces plant hormones that benefit plant development and enhances molecular responses to biotic and abiotic stress (Contreras-Cornejo et al., 2016). Formulations based on T .asperellum, T. atroviridae, T. virens and T. harzianum are commercially available and used in agriculture (Woo et al., 2014).
Production of toxic secondary compounds from distinct metabolic pathways produced by Trichoderma spp. contribute to suppress the growth of neighbor phytopathogenic fungi at the soil or rhizosphere (Keswani et al., 2014). Mycoparasitism is another biological feature of Trichoderma that influences fungal population in soil and rhizosphere. Trichoderma can form hook-like structure attached to hyphae of other fungi referred to as ‘prey’. Physical pressure derived from the hook-like structures and the activity of cell wall degrading allows acquisition of carbon and nitrogen sources from the prey (Harman et al., 2004).
T. harzianum (teleomorph: Hypocrea lixii) can colonize the epidermis and the outer cortex of cucumber roots (Yedidia et al. 2001). While interacting with roots, Trichoderma stimulates the execution of the induced systemic resistance (ISR) that strengthen the defense responses (Martínez-Medina et al., 2013)jasmonic acid (JA. Besides the antagonistic activity, Trichoderma spp. produce hormones such as auxins that boost plant growth (Contreras-Cornejo et al. 2009).
Due to the need to sustain the production of banana fruits in Mexico, it is necessary to prospecting biocontrol agents, native from the crop area. Here, we report the identification of a potential biocontrol agent isolated from pseudostems of “platano macho” (Musa sp. AAB) grown in the coast of Oaxaca. Sequencing of the internal transcribed spacer (ITS), large subunit ribosomal DNA (LSU) and the translation elongation factor-1α (TEF-1α) confirmed the specie as T. harzianum. The results validated its beneficial effects and suggest that the exploration of plant-associated microbial communities is a reliable first step to develop novel products of crop protection.
Materials and methods
Isolation and identification of a novel Trichoderma strain
The strain was isolated from fragments of pseudostems coming from ‘plátano macho’ (Musa sp. AAB cv. Macho) at the municipality of Villa de Tututepec de Melchor Ocampo, Oaxaca, Mexico (16° 08′N, 97° 36′ W) in parallel of the experiments of isolation of Fusarium spp. previously reported (Maldonado-Bonilla et al., 2019). Cut and washed tissue samples of approximately 0.5 × 0.5 cm were disinfected with 70% ethanol for 1 min, 3% sodium hypochlorite for 10 min, and then, were rinsed three times with sterile distilled water. The pieces of disinfected plant tissue were placed in Potato Dextrose Agar (PDA) plates and incubated for 1 week at room temperature. A fast growing mycelium of white color coming from one of the pieces were transferred to fresh PDA plates, after 1 week, mycelium turns dark green and a concentric ring is formed due to the production of conidia. This fungus was denominated M110, and after identification it was consequently referred to as ThM110. Conidia from one PDA plate were harvested with glycerol 50% to store at -80 °C. Morphology was visualized by optical microscopy and lactophenol-cotton blue staining.
Polymerase chain reaction
Extraction of genomic DNA from ThM110 was performed following the protocol reported to isolate DNA of Fusarium spp. (Lin et al., 2009). After extraction, DNA was diluted and subject to PCR by using MyTaqR (Bioline). Amplification of ITS was performed by using the universal primer combination ITS1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) under the following conditions: 5 min at 95 °C of initial denaturation; 35 cycles of 94 °C for 30 s (denaturation), 55 °C for 60 s (annealing) and 72 °C for 60 s (extension); final extension of ٥ min at ٧٢ °C. Amplification of a LSU fragment was performed with the primer combination LROR (5’-ACCCGCTGAACTTAAGC-3’) and LR5 (5’- TCCTGAGGGAAACTTCG-3’) by the following protocol: 5 min at 95 °C of initial denaturation; 35 cycles of 94 °C for 60 s (denaturation), 55 °C for 60s (annealing) and 72 °C for 60 s (extension); final extension of 5 min at 72 °C. The amplification of the TEF-1α fragment was performed with the primer combination tef71f (5’-CAAAATGGGTAAGGAGGASAAGAG-3’) and tef997R (5’- CAGTACCGGCRGCRATRATSAG-3’). The ‘touchdown’ amplification program was 4 min of initial denaturation at 94 °C; 4 cycles each of 60 s at 94 °C (denaturation), 90 s at 70 °C (annealing) and 90 s at 72 °C (extension); followed by 26 cycles with annealing temperature decreasing by 0.5 °C per cycle from 68 °C to 55 °C, followed by 12 cycles with annealing at 55 °C; and with a final extension of 7 min at 72 °C. These PCR conditions are available in the website of the International Subcommission on Trichoderma and Hypocrea Taxonomy (http://www.isth.info/isth/methods/method.php?method_id=9) and has been validated at least in another study (Jiang et al. 2016)Ascomycota. Amplification of PCR products was confirmed by agarose gel electrophoresis stained with Eco-Stain (Biobasic).
Cloning of TEF-1α
We decided to clone the TEF-1α amplicon since it is longer than 900bp which could hinder the full sequencing by using the above mentioned tef71f and tef997R. After the amplification, the PCR product was cloned into pGEM T-Easy (Promega) following the manufacturer’s instructions. Ligation products were used to transform chemically competent E. coli JM109 cells. The transformation reactions were platted out in LB agar supplemented with 100 mg mL-1 ampicillin, 10 µL of 100mM IPTG (Isopropyl β-D-1-thiogalactopyranoside) and 50 µL of 20 mg µL-1 of X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside). Plasmids of white colonies were extracted and digested with EcoRI to screen for clones carrying the TEF-1α PCR product. The plasmid of the selected colony (pGEM-TEF-1α) was purified with the QIAPrep Spin Miniprep Kit (QIAGEN).
DNA sequencing and analysis
The amplicons of the ITS and LSU regions were directly sequenced by capillary electrophoresis using the primers mentioned for the PCR. The purified pGEM- TEF-1α was sequenced by capillary electrophoresis with the primers T7promoter (5’-TAATACGACTCACTATAGGG-3’) and SP6 (5’- ATTTAGGTGACACTATAG-3’), both of them are suitable for the sequencing of DNA cloned into pGEM-T-Easy. Sequencing service was provided by Macrogen (https://dna.macrogen.com/eng/index.jsp). After the assembly of the full sequences, they were used as queries to search for nearest matches in Genebank using the BLASTN algorithm optimized for highly similar sequences in the nucleotide collection database (nr/nt) of the National Center for Biotechnology Information (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences were considered conspecific when the percentage of identity was higher than 97% between a given record and our queries (Ávila-Díaz et al., 2013). The sequences of ITS, LSU and TEF-1α were submitted to Genebank with the accession numbers MN555334.1, MT549065.1 and MN557854.1, respectively.
In vitro antagonistic activity
In vitro confrontation assays were performed to get a first insight into the antifungal capabilities of M110. The growth of the wilting-causing strains M1a, M5, M7, M104 and M108 of Fusarium spp. (Maldonado-Bonilla et al., 2019)a sampling was performed in symptomatic plantains growing in fields of Oaxaca, a coastal southern state of Mexico. A phylogenetic analysis based on the sequences of TEF 1-α and IGS revealed that three isolates belonged to the Fusarium oxysporum species complex, while two other isolates were identified as members of the Fusarium fujikuroi species complex. Furthermore, isolates from the same complex shared the same ITS2 sequence. Inoculation using spores of each isolate on the roots of Musa sp. AAB cv. Manzano produced wilting symptoms of varying severity, suggesting that the Fusarium wilt might not be caused only by Fusarium oxysporum f. sp. cubense. PCR-based detection of Secreted in Xylem (SIX was determined in the presence of M110. Fusarium spp. and M110 spores were spread in PDA plates and incubated at 28°C for one week. After this period, the mycelia cover the whole plate. Squares of 0.5 cm2 of newly grown mycelium from each phytopathogen were inoculated close to the margin of a 10 cm diameter Petri dish with PDA. At the opposite margin of the same Petri dish, a similar square of newly grown mycelium of M110 was inoculated. A Petri dish only inoculated with the phytopathogens were used as negative controls. The growth of phytopathogens was recorded at 7 days after inoculation. Every assay included three technical replicates and was repeated three times. The percent of growth was calculated by dividing the growth of the phytopathogen in presence of ThM110 by the growth of the phytopathogen alone (Askew and Laing, 1994).
Assessment of plant protection
Musa sp. AAB cv. ‘manzano’ (‘plátano manzano’) plants used for this assay were developed by following the in vitro micropropagation method established in our workgroup (Maldonado-Bonilla et al., 2019)a sampling was performed in symptomatic plantains growing in fields of Oaxaca, a coastal southern state of Mexico. A phylogenetic analysis based on the sequences of TEF 1-α and IGS revealed that three isolates belonged to the Fusarium oxysporum species complex, while two other isolates were identified as members of the Fusarium fujikuroi species complex. Furthermore, isolates from the same complex shared the same ITS2 sequence. Inoculation using spores of each isolate on the roots of Musa sp. AAB cv. Manzano produced wilting symptoms of varying severity, suggesting that the Fusarium wilt might not be caused only by Fusarium oxysporum f. sp. cubense. PCR-based detection of Secreted in Xylem (SIX. In vitro rooted plants produced by this method were transferred into covered plastic containers with vermiculite rinsed with 25% MS solution without sucrose, and incubated at 26 °C with a 16 h light and 8 h darkness photoperiod. The covers of the containers were gradually removed to prevent dryness. After three weeks of incubation, the plants already had cuticle and produced two newly emerging leaves. Then, the plants were taken from the plastic container. The two older leaves formed during the in vitro micropropagation were excised, and the roots were thoroughly washed with water to eliminate the vermiculite. In parallel, the M110 conidia were harvested with sterile distilled water from a Petri dish with one week-old cultures grown in PDA. The suspension was adjusted to 106 conidia mL-1. Twenty plants were inoculated with M110 by submerging the roots into the conidia suspension for 1 min. Additional 20 plants were mock-treated with water. Later on, 10 plants inoculated with M110 and 10 mock-treated plants were infected by adding 10 mL of a Fusarium sp spores suspension. M5 adjusted to 106 conidia mL-1. The growth of Fusarium sp. M5 and preparation of spore suspension was performed as mentioned above for M110. Out the remaining plants, 10 were reference plants inoculated with M110 and 10 plants only inoculated with water were kept as additional testing conditions. Symptoms were recorded 10 weeks after inoculation by sorting plants into 4 categories: Healthy, yellow leaves, yellow leaves with partial necrosis and necrosis in whole leaves (Maldonado-Bonilla et al., 2019). The experiment was repeated twice.
Results and discussion
T. harzianum is a cosmopolitan fungus found as saprophyte in soils, rotting plants, wood, bark as well as associated with living organisms such as plants and other fungi (Chaverri et al., 2015). A close interaction between T. harzianum strain T-2013 and cucumber roots induces defense responses, solubilizes phosphate and increases its concentration in roots and shoots, which in turns enhances the plant growth (Yedidia et al., 2001). The detection of different T. harzianum strains in roots, stems, leaves and plumules of cacao plants suggests its adaptation to set an endophytic growth and a tight interaction with plants (Bailey et al., 2008). Furthermore, T. harzianum as one of the species most commonly isolated as endophytes in the sapwood of tropical trees (Almeida et al., 2018), which might be an indirect evidence of translocation in planta. Such conserved feature of this species might be the crucial factor to isolate M110 from living pseudotems of ‘plátano macho’.
Lactophenol-cotton blue staining was performed in order to identify the M110 species. Trichoderma harzianum M110 is recognized in PDA culture medium for its rapid growth and its yellow-green coloration with concentric growth rings formed by the production of conidia. Microscopically, it is characterized by erect, hyaline conidiophores with bottle-shaped phialides (5.5 -) 6-6.8 (-8.5) µm in length and (1.5-) 1.6-1.7 (-2.8) µm in the upper part (Figure 1A). The conidia are hyaline to slightly greenish in KOH (5%), subglobose to slightly oblong, 2.4-2.9 (-3.3) X (2.0-) 2.3-2.7 µm, with a smooth wall (Figure 1B). The description agrees with that made by Gamms and Bissett (2002). This morphology suggested that the isolated specie is T. harzianum.
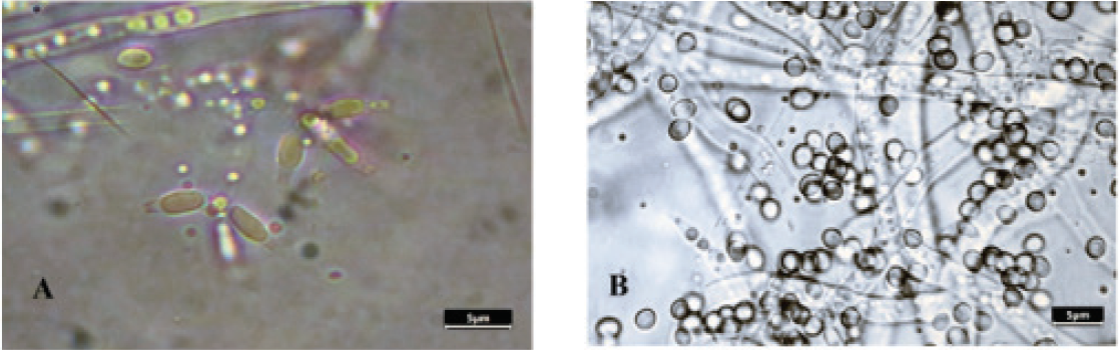
Figure 1 Optical microscopy reveals that M110 has characteristic features of T.
harzianum. A) Bottle-shaped phialides. B) Subglobose to
slightly oblong conidia. Bar: 5 mm.
Figure 1. Microscopía óptica revela que la cepa M110 tiene
características típicas de T. harzianum. A) Fiálides
con forma de botella. B) Conidios subglobosos a ligeramente oblongos.
Barra: 5 mm.
T. harzianum and closely-related species such as T. guizhouense, T. lixii, and T. inhamatum form the T. harzianum species complex (THSC). It is proposed that every species of the THSC has a unique geographic distribution, for example T. harzianum is distributed in North America and Europe, while T. inhamatum is distributed in South America. In this context, the location of the isolation of M110 (the Pacific coastline of Mexico) agrees with the expected distribution of this species (Chaverri et al., 2015).
A bias in the distribution of the species of the THSC might be influenced by the climate conditions, soil composition and the plant diversity that can allow of the endophytic growth. Members of THSC share a quite similar morphological characters, thus, additional criteria such as sequencing of phylogenetically informative loci are necessary to define the species. The ITS is considered the fungal barcode sequence (Ratnasingham and Hebert 2007). The full ITS sequence is composed by the 5.8S ribosomal gene flanked by the region ITS1 at the 5’ and the ITS2 at the 3’(Pryce et al., 2003). ITS1 and ITS2 are transcribed together with the rest ribosomal operon including the 18S (SSU), 5.8S and the 28S (LSU) rRNAs. During maturation of rRNAs, the ITS1 and ITS2 are degraded. The LSU sequence is used as a marker for fungal molecular taxonomy, but, due to the lack of evolutionary pressure, most of the polymorphisms are located into the ITS1 and ITS2.
The full ITS and a fragment derived from the LSU were amplified by PCR and sequenced. Both sequences were submitted as query in Genebank to search for highly similar sequences by using the BLASTN algorithm. Table 1 shows the two matches of highest identity. The values of percent identity above 99% suggest the identified species is T. harzianum. In order to discriminate between members of the THSC and confirm the identity of M110, a fragment of the TEF-1α gene was amplified, cloned and analyzed. The sequenced TEF-1α fragment of M110 was subject to the workflow used to analyze ITS and LSU. The results are presented in Table 1. The best matches correspond to collection strains T. harzianum, thus, M110 is a novel strain of T. harzianum isolated from pseudostems of banana. To our knowledge, this is the first report is this species associated to banana plants in Mexico.
Table 1: Identity of the full length sequence ITS, and the derived fragments from LSU and TEF-1α
of M110. The first two sequences of highest percent identity and
coverage are shown. The access numbers of the corresponding sequences
are indicated at the far right column.
Tabla 1. Identidad de la secuencia completa ITS y de los
fragmentos derivados de LSU y TEF-1α de la cepa M110. Se muestras las
primeras 2 secuencias con mayor porcentaje de identidad y cobertura. Los
números de acceso de las correspondientes secuencias se indican en la
última columna de la derecha.
| Sequence | % Identity | % Coverage | E-value | Closest species match | Accession number |
| ITS | 99.68 | 100 | 0 | T. harzianum NECC30498 | MH153641.1 |
| 99.68 | 100 | 0 | T. harzianum AA1I10F1 | KX421472.1 | |
| LSU | 99.88 | 100 | 0 | T. harzianum CBS 130681 | MH877292.1 |
| 99.88 | 100 | 0 | T. harzianum CBS 226.95 | MH874152.1 | |
| TEF-1α | 97.30 | 93 | 0 | T. harzianum (H. lixii) CIB T131 | EU279988.1 |
| 96.14 | 94 | 0 | T. harzianum JBT1244 | AY605784.1 |
We performed confrontation assays in order to demonstrate its potential antagonistic activity. M110 and 5 strains of phytopathogenic Fusarium spp. were co-inoculated in PDA. Negative controls of PDA plates solely inoculated with the phytopathogens were run in parallel. After 4 days, when fungi start to touch each other, the growth of Fusarium spp slightly decrease in the presence of M110 (data not shown), and the difference is enhanced at 7 days post inoculation when M110 covers the plate and the growth of Fusarium spp. is fully arrested. The results of representative plates after 7 days of inoculation are shown in Figure 2. Table 2 shows the results of Fusarium spp growth percentage in the presence of M110. A scale of 5 classes based on the phytopathogen growth percentage with Trichoderma respect to the phytopathogen growth alone is recommended to rank the antagonism (Askew and Laing, 1994). The Class 1 is the most effective and does not allow the growth of the phytopathogen. In Class 2, the pathogen growth is up to 25%, in Class 3, the pathogen growth reaches up to 50%. In Class 4, both fungi locked at the point of contact; and Class 5 indicates the phytopathogen entirely grew in the medium, inhibiting the Trichoderma growth. The results of the Fusarium spp. growth percentage in the presence of M110 are shown in Table 2. The interactions M1a vs, M110, M5 vs M110, M7 vs. M110 and M108 vs. M110 displayed quite consistent results below 50% of growth. The interaction M104 vs. ThM110 gave high standard deviation, but the inhibition is visually evident. Therefore, M110 is associated to the Class 3 of antagonism.
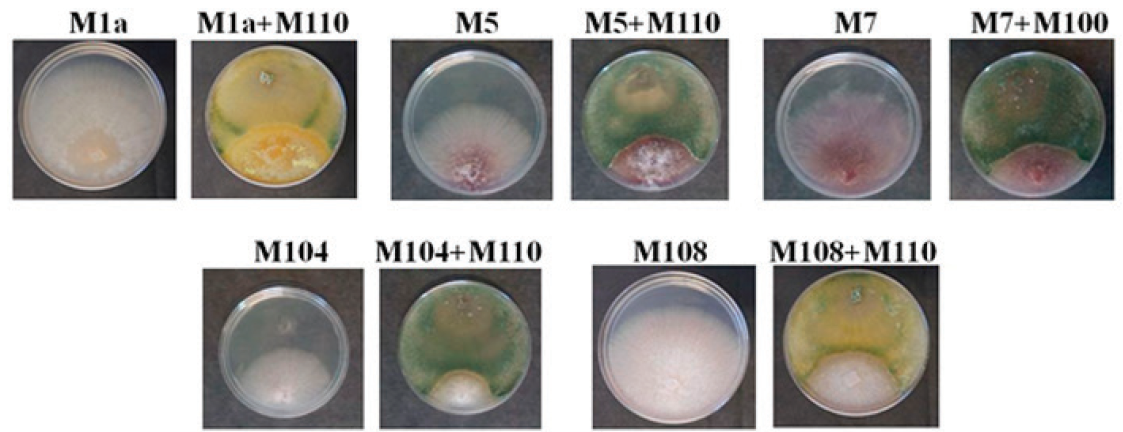
Figure 2 Representative photographs of the confrontation assays Fusarium spp. vs. T.
harzianum M110 show that the mycelial growth of Fusarium
spp. is inhibited by M110. Plates inoculated only with Fusarium were
used as controls. The name of the inoculated strains is indicated above
each plate. Photographs were taken 7 days post inoculation.
Figura 2. Fotos representativas de ensayos de confrontación
Fuarium spp. vs. T. harzianum M110 muestran que el
crecimiento micelial de las cepas de Fusarium spp. se inhibe en
presencia de M110. Placas inoculadas sólo con Fusarium se usaron como
controles. El nombre de las cepas inoculadas se indica sobre cada placa.
Las fotografías se tomaron 7 días después de la inoculación.
Table 2 Percent of mycelial growth of Fusarium spp. in PDA plates co-inoculated with M110
respect to mycelial growth of Fusarium spp. in PDA plates without M110.
See material and methods for details.
Tabla 2.
Porcentaje de crecimiento micelial de Fusarium spp. en placas de PDA
co-inoculadas con la cepa M110 respecto al crecimiento micelial de
Fusarium spp. en placas PDA sin M110. Para detalles, ver materiales y
métodos.
| Fusarium strain | Percent of growth (DAY 7) |
| M1a | 34.85 + 5.52 |
| M5 | 37.46 + 4.20 |
| M7 | 33.14 + 5.49 |
| M104 | 53.05 + 12.93 |
| M108 | 29.56 + 5.26 |
Infection assays in the presence or absence of M110 were performed to validate the ability of M110 to antagonize causal agents of Panama disease in the soil or rhizosphere. After the Foc spore germinate, nascent hyphae adhere to the root epidermis, penetrate, and invade apoplast. When xylem vessels are reached, the fungus is dispersed throughout plant, causing discoloration of the rhizome and yellowing of leaves, and later on browning of the vascular bundles, necrosis and splitting of the pseudostem (Ploetz, 2015). Under our experimental conditions, strain M5 displays the more severe symptoms towards Musa sp. AAB cv ‘manzano’. A M5 spores suspension was inoculated in roots of mock plants and plants previously inoculated with M110. The M110 potential protective effect is shown in Figure 3. Strain M5 causes severe wilting of the leaves, but the symptoms notably decreased in plants pre-inoculated with M110 (Figure 3A). By comparing M110-inoculated and control plants, we suggest that M110 does not cause visual negative effects to Musa sp. AAB ‘manzano’. Figure 3B shows the distribution of symptoms in the plants grown under these experimental conditions and confirms the pre-inoculation of M110 decreases of wilting of leaves caused by M5. Under these conditions, no further symptoms were detected. Therefore, M110 seems to be innocuous to plants and its inoculation into roots decreases the Panama disease symptoms, possibly by the antagonism characteristic of the Trichoderma genus.
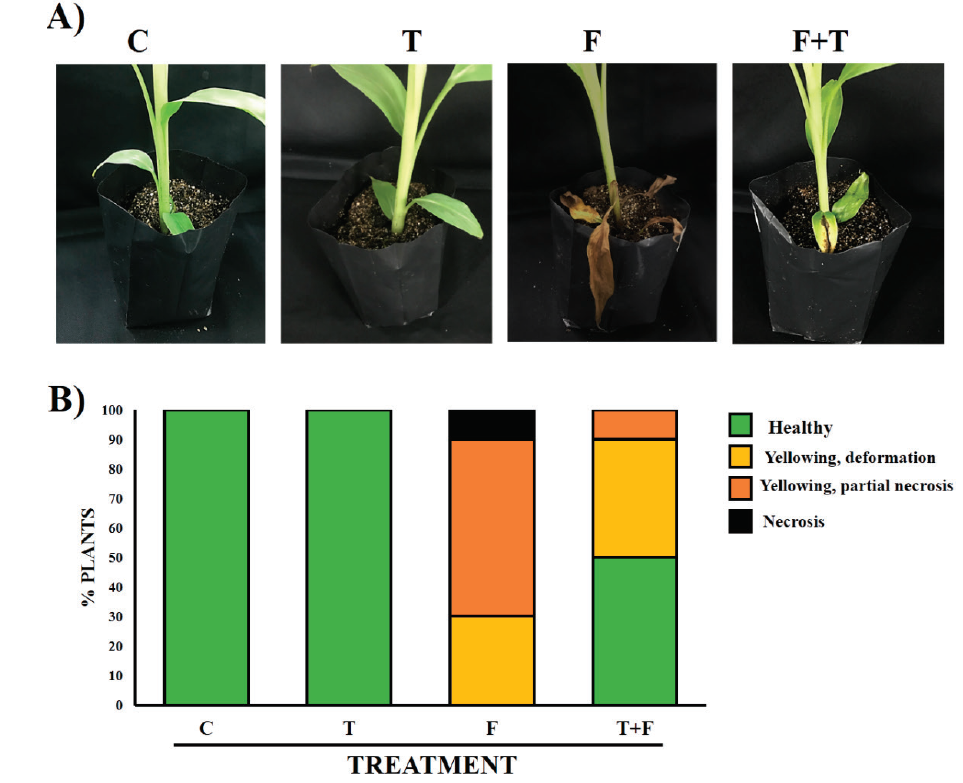
Figure 3 Evidences of the protective effect of M110 against Fusarium sp. M5. Spores solutions of
M110 and M5 were inoculated in roots of Musa sp. AAB cv. ‘manzano’. A)
The effect is visible in plants 10 weeks after inoculation. B) Plants
were classified according to the displayed symptoms of different
severity at 10 weeks after inoculation. C, non-inoculated control. T,
T. harzianum M110. F, Fusarium sp. M5. F+T,
T. harzianum M110 + Fusarium sp. M5. See materials
and methods for details of the inoculation procedure.
Figura 3. Evidencias del efecto protector de
M110 contra Fusarium sp. M5. Soluciones de esporas de M110 y M5 se
inocularon en raíces de Musa sp. AAB. cv ‘manzano’. A) Efecto en plantas
es visible a 10 semanas después de inocular. B) Las plantas fueron
clasificadas de acuerdo a los síntomas mostrados 10 semanas después de
la inoculación. C, plantas control no inoculadas, T, plantas inoculadas
con T. harzianum M110. F, plantas inoculadas con
Fusarium sp. M5. F+T, T. harzianum M110 + Fusarium sp.
M5. Ver materiales y métodos para detalles de procedimiento de
inoculación.
Antifungal compounds such as peptabiols, polyketides and terpenes are produced by Trichoderma spp (Keswani et al., 2014). Such metabolites might limit the proliferation of other fungi that compete for carbon and nitrogen source and represent a benefit to the plant when phytopathogens are inhibited. Remodeling of the cell wall by the activity of chitinases, glucanases and proteases is required for hyphal growth and branching (Verdín et al., 2019). Those extracellular enzymes can reach the hyphae of neighbor fungi and restrict their growth. Coiling around fungal host enhances the antagonistic effect of Trichoderma (Guzmán-Guzmán et al., 2018), since physical proximity between Trichoderma and host facilitates the effect of antifungal metabolites and the lytic enzymes mentioned above. Therefore, diffusible factors produced by T. harzianum M110 and the coiling might contribute to the in vitro growth arrest of Fusarium spp., and, in the same way, they could obstruct the development of Fusarium sp. M5 in the rhizosphere of Musa sp. AAB ‘manzano’.
Besides the antagonism, Trichoderma might elicit mechanisms of protection. Genes encoding biosynthetic enzymes of biotic stress-related plant hormones such as salicylic acid (SA) and jasmonic acid (JA), have been identified in the T. atroviridae genome (Guzmán-Guzmán et al., 2018). A number of Trichoderma-derived proteins and non-ribosomal peptides can also trigger defense responses such as production of phytoalexins, reactive oxygen species, or induction of genes encoding pathogenesis-related proteins (Ramírez-Valdespino et al., 2019). These reactions have to be explored in the future in order to have a better understanding about the protective effect of M110 in ‘plátano manzano’. Both SA and JA-dependent signal pathways are stimulated by T. virens in tomato plants, and induced resistance against Fusarium oxysporum f.sp. lycopersici (Fol) (Jogaiah et al., 2018). As the pathogenicity mechanisms of Foc and Fol are similar, we have to consider the role of the SA and JA pathways in banana, either elicited by hormones produced by the fungus or by molecules that trigger hormone biosynthesis and signaling in the plant. These possibilities can be investigated in M110 and banana plants.
A combination of T. harzianum and Glomus spp. suppressed the infection of FocTR4 towards the ‘Lakatan’ bananas, a top-selling cultivar in Philippines (Castillo et al., 2019). A combined formulation of T. harzianum and Bacillus subtilis enhances the uptake of minerals and biomass production in swiss chard (Venegas-Gonzalez et al., 2019). That illustrates additional effects of T. harzianum that might be investigated in the future for the interaction between M110 and banana plants.
We report here a T. harzianum strain isolated from plants growing in southern Mexico, which has features of a biocontrol agent, and it can be efficient to protect plants against Panama disease. Some banana producers have faced disease-related restrictions and bans due to Panama disease (FAO, 2017). Mexico has an important trade to export bananas to China (Gobierno de México, 2020) thus, it is necessary to be alert in securing the plant health and production of pathogen-free bananas.
Conclusions
Due to the economic importance of banana in Mexico, and the need to sustain the production of fruits, it is necessary to prospecting biocontrol agents native from the soil of the crop area and adapted to interact with cultivars grown in Mexico. Here, we demonstrated the antagonism of T. harzianum M110 towards Fusarium spp., and demonstrated the potential of this novel strain as biocontrol agent. These findings encourage us to investigate the molecular basis of its antagonism and interaction with plants. Communication between microbiologists, phytopathologists and biotechnologists is necessary to develop large-scale biopesticide products as alternative to protect plants against Panama disease.











 nueva página del texto (beta)
nueva página del texto (beta)


