Introduction
Soil is the most important biological matrix on Earth, where a broad microbial diversity is developed including prokaryotes, eukaryotes and virus. Microbiota play important roles in the edaphogenesis process, biogeochemical cycles, xenobiotics degradation (herbicides, insecticides and hydrocarbons), but above all in plant growth, carried out by the so called plant-growth promoting bacteria (PGPB) (Hillel, 1998; Jaramillo et al., 1994; Tarbuck et al., 2005). The PGPBs can be free-living or symbiotic relationship, they have mainly isolated from grasses soils, such as corn (Loredo-Osti et al., 2004). It is estimated that microbial biodiversity in soil ecosystems is mainly represented by eukaryotic organisms. For example, one gram of rhizospheric soil can contain up to 10 billion microorganisms and more than 30,000 prokaryotes species (Egamberdieva et al., 2008; Mendes et al., 2011). Soil microorganism study not only lies in ascertaining their importance in product generation, metabolic processes and biotechnology capacities, but also in their direct relationship with nutrient utilization. A few years ago, soil bioprospecting only consisted of microorganism culture through traditional microbiological techniques (Handelsman et al., 2002; Torsvik and Øvreås, 2002). However, these culture methods only allow a recovery of between 0.1-10% total microorganisms (Escalante-Lozada et al., 2004). This can be explained because the nutrient requirements of all microorganisms are unknown. In addition, the precise physicochemical conditions of their natural environment have not been documented, as well as the information on the symbiotic, commensal or parasitic relationships that are maintained in a microbial community. For this reason, soil ecosystems are largely unknown (Keller and Zengler, 2004; Zengler et al., 2002). Schloss and Handelsman (2003) found that most of strains in the soil belong to four phyla: Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria, which represent 20% of the bacterial community in the soil.
An alternative to microorganism recovery related to the degradation of some metabolite or substrate is culture enrichment. Enrichment can be of two types: in the laboratory adding some specific substrate or media and by in situ enrichment before isolation (Sar and Islam, 2012). Thus, diverse microorganisms, metabolic pathways and enzymes related to the degradation of complex carbohydrates (chitin, lignocellulosic residues) and simple, fats and oils among others have been able to be identified (Beloqui et al., 2009; Jacquiod et al., 2013; Peña-García et al., 2016; Wang et al., 2016).
Sugarcane is a perennial grass grown in many tropical countries. Globally, it is one of the most important staple crops, both in terms of total production (ranked #1 at 1,685 million tons) and area cultivated (#13 at 23.8 million ha) (2010 data; http://faostat.fao.org). In many tropical countries, sugarcane production represents the most important land-use and agricultural commodity; in countries such as Brazil, its importance is due to ethanol production.
The Papaloapan Basin is located in southeast Mexico. It is the second most important hydric region of Mexico with warm humid weather. Its soils are so fertile that sugarcane is the main crop harvest in the region with 30% coverage (OEIDRUS, 2005). Main soil types are ferrosol, fluvisol and phaeozems, which have particular characteristics that allow the development of the microorganisms (CODICE, 2010). Until now, diversity studies using enriched cultures in soils sown with sugarcane in the Papaloapan Basin (a megadiverse zone) have not been carried out. The objective of this research was to obtain a preliminary estimate of soil biodiversity related to starch and cellulose hydrolysis in soils sown with sugarcane in the Papaloapan Basin via enriched cultures. The results represent a preliminary estimate of the microbiota present and to associate them with crop type, in order to generate useful knowledge for future research or for the industries and institutions related to sugarcane culture and biofuel production from lignocellulosic residues.
Method
Soil sampling protocol
Samples were collected (April 2013) from four different sugarcane sites (CP 72-2086 and Mex 69-290 varieties) in rhizospheric soil, belonging to the same plot (lat. 18.00˚N, 96.07˚W and 46 m.a.s.l.). Vegetable matter was removed, and the samples were placed in plastic bags and stored at 4°C until its use and chemical analysis.
Soil physico-chemical characterization
All soil samples collected were mixed and an aliquot was characterized. Saturated soil paste extract pH, electrical conductivity (EC) of soluble ions (dSm-1) (Jenway Model 4520 Laboratory Conductivity/TDS Meter), saturation percent (SP), cations (Na+, K+, Mg2+, Ca2+) and anions (SO42-, Cl-, HCO3-, CO32-) were determined for each sample (Jackson, 1973). Soil calcium carbonate was measured by the calcimeter method (Nelson, 1982). Mechanical analysis was carried out by the international pipette method (Piper, 1966), and the textural class was ascertained from SP and confirmed by textural triangle.
Enriched cultures
Soil samples were homogenized, and 2 g soil sample were placed on 200 mL Luria-Bertani medium alone (Sigma-Aldrich), with carboximetilcellulose (Sigma-Aldrich) (LB-CMC-1%, W/V), and starch (Jalmek) (LB-starch 1%, W/V). Liquid cultures were maintained at 37°C, 180 rpm for 48 h. Subsequently, biomass was collected by centrifugation at 5000 rpm (Eppendorf 5418R) and stored at -20°C until its analysis.
DNA extraction and amplification of 16S rDNA gene
From the collected biomass, metagenomic DNA was extracted using Microbial DNA Isolation Kit (MOBIO) following the instructions of the supplier. A polymerase chain reaction (PCR) assay was designed to amplify a 1.6 kb region of the 16S gene. For the PCR, 35-50 ng extracted DNA were used with a total volume of 50 μL using 0.5 - 1 μL (10 mM) of the fD1 primers (5’-CCG AAT TCG TCG ACA ACA GAG TTT GAT CCT GGC TCA G-3’) and rD1 (5’-CCC GGG ATC CAA GCT TAA GGA GGT GAT CCA GCC-3’), 1.4 μL (5 U/μL) of Platinum Taq DNA polymerase (Invitrogen, cat. 10966-026) and 3 mM MgCl2 under the following conditions: 3 min at 94˚C, 30 cycles of 30 seconds at 94˚C, 30 seconds at 56˚C, and 2 min at 72˚C, plus an additional 5 min cycle at 72˚C (Gutiérrez-Lucas et al., 2014). PCR products were analyzed on 1% agarose gels.
Purification and cloning of the PCR products
PCR products were purified using a GeneJET PCR Purification kit (Thermo Scientific, cat. K0701, USA) following the indications of the supplier. PCR products were cloned on the pCR-XL-TOPO vector of the TOPO XL PCR Cloning kit (Invitrogen, cat. K4700-20, USA) according to supplier specifications, (80 ng/μl DNA, 10 ng/μl vector). PCR products were analyzed by sequencing (Macrogen Inc, South Korea).
OTUs and Phylogenetic analysis
Operational taxonomic units (OTUs) and diversity indexes were obtained with the typically implemented mothur software pipeline (Schloss et al., 2009). The program was run with the 35 obtained sequences. No chimeric sequences were found through the “chimera.uchime” algorithm incorporated in the platform. A distance matrix was built using "dnadist", included in PHYLIP software (v.3.69 Felsenstein, USA, 2005) and, with the cluster command included in mothur; the sequences were assigned to 20 OTU’s. The cluster method used the “furthest neighbor” in which all the sequences within an OTU are at the most 0.03 distant (or 97% similar) from all of the other sequences with the OTU. After clustering, the sampling effort was evaluated by a rarefaction curve and the diversity among the sequences was calculated with the Chao1, Shannon and Simpson indexes (Figure 6).
Subsequently, sequence homology and analysis were performed using a BLASTn tool program available on-line (ncbi.nlm.nih.gov); the sequences of the species of the resulting genus were obtained (img.jgi.doe.gov) and aligned with the MUSCLE program (ebi.ac.uk/Tools/msa/muscle/) to obtain a profile. With the alignment of the profile, a search was made for the best method of phylogenetic inference with the FindModel program (hiv.lanl.gov). For all inferences, the maximum likelihood method was GRT + G (General Time Reversible plus Gamma distribution). The sequences obtained from the isolates were aligned with the profile obtained from GenBank with the Clustal_X program in the profile alignment mode. After the final alignment, phylogenetic inference was carried out with the above-mentioned method and 500 bootstrap replicates with the program MEGA6 program. The phylogenetic tree was constructed using the closest genera.
Results and discussion
Soil physico-chemical characteristics
Analyzed soil was composed of 51.96% clay, 30% silt and 18.04% sand; its textural class was clay, with 2.06 g/cm3 density, pH almost neutral (6.32) and the EC value was 0.147 dSm-1. Organic material and nitrogen content were moderately high (Table 1). Analyzed soil satisfied the physico-chemical conditions required for sugarcane harvest and showed similar parameters to Brazilian soil sown with this crop (Rachid et al., 2012; Rachid et al., 2016). Therefore, these data could be used as reference in sugarcane production in Mexico.
Table 1 Chemical characterization of the soil sample from the studied area.
| Parameter | Result | Unit | Observations |
|---|---|---|---|
| Organic material | 3.09 | % | Moderately high |
| Total Nitrogen | 0.177 | % | Moderately high |
| Assimilable Phosphate | 6.20 | mg.kg-1 | Low |
| Ca+2 | 7216.80 | mg.kg-1 | High |
| Mg+2 | 304.88 | mg.kg-1 | Medium |
| Na+ | 44.85 | mg.kg-1 | Very low |
| K+ | 149.37 | mg.kg-1 | Moderately low |
| Fe | 38.70 | mg.kg-1 | High |
| Cu | 2.67 | mg.kg-1 | High |
| Zn | 1.26 | mg.kg-1 | Moderately low |
| Mn | 20.96 | mg.kg-1 | Moderately high |
| B | 0.35 | mg.kg-1 | Very High |
| CIC (Cationic Interchange Capacity) | 36.80 | cmol.kg-1 | Moderately high |
Diversity analysis
In this research, 35 OTUs were obtained 33 of which were identified from the LB-starch library, two OTUs from the LB-CMC library and none from the LB library. Sequence analysis indicates that 17 OTUs are related to Clostridium, nine to Bacillus, six to Enterococcus, two to Lysinibacillus (one in LB-starch and the another in LB-CMC libraries) and one to Citrobacter (found exclusively in LB-CMC library) (Table 2). In a previous study carried out in soil sown with sugarcane in the Papaloapan Basin, 12 and 6 strains with amylase and cellulase activity respectively were isolated and identified by biochemical and molecular tests (16S rDNA gene sequencing). Isolated strains were related to Bacillus (phylum Firmicutes, class Bacilli), Arthrobacter (phylum Actinobacteria, class Actinobacteria) and Pseudomonas (phylum Proteobacteria, class Gammaproteobacteria). Some of these could be new species of certain genera, due to the low similarity they showed with the closest strains (Montor et al., 2011).
Table 2 OTUs obtained from enrichment media with starch and cellulose.
| Culture media | OTU | Genera | ID_NCBI | Best-hit/coverage |
|---|---|---|---|---|
| LB-1% starch | ANC3 | Bacillus | CP020002.1 | 100/99 |
| ANC7 | Bacillus | CP020002.1 | 100/99 | |
| ANC8 | Enterococcus | CP019512.1 | 100/99 | |
| ANC9 | Clostridium | KM497177.1 | 98/99 | |
| ANC14 | Clostridium | EU773775.1 | 92/96 | |
| ANC15 | Enterococcus/Unc | EU723856.1 | 97/91 | |
| ANC16 | Bacillus | CP019726.1 | 100/99 | |
| ANC17 | Clostridium | X73446.1 | 99/94 | |
| ANC18 | Clostridium/Unc | HG917272.1 | 100/94 | |
| ANC19 | Clostridium | CP010994.1 | 100/99 | |
| ANC22 | Lysinibacillus | KY643638.1 | 100/99 | |
| ANC23 | Bacillus | KP209387.1 | 100/97 | |
| ANC26 | Bacillus | CP018935.1 | 100/99 | |
| ANC28 | Clostridium | AB161374.1 | 100/99 | |
| ANC30 | Clostridium | AB161369.1 | 100/99 | |
| ANC34 | Clostridium | AB161372.1 | 99/99 | |
| ANC35 | Enterococcus | CP003726.1 | 100/96 | |
| ANC36 | Clostridium | AB161372.1 | 100/98 | |
| ANC42 | Clostridium | EU775706.1 | 92/97 | |
| ANC43 | Bacillus | CP014486.1 | 100/99 | |
| ANC46 | Clostridium | AP017630.1 | 100/99 | |
| ANC50 | Bacillus | CP018935.1 | 100/100 | |
| ANC52 | Clostridium | JF428988.1 | 99/94 | |
| ANC53 | Clostridium | KC835992.1 | 99/96 | |
| ANC55 | Clostridium/Unc | HG917272.1 | 100/95 | |
| ANC60 | Clostridium | CP010994.1 | 100/93 | |
| ANC61 | Enterococcus | JF772098.1 | 99/97 | |
| ANC65 | Clostridium | CP010994.1 | 100/99 | |
| ANC67 | Enterococcus | CP019512.1 | 100/99 | |
| ANC68 | Enterococcus | CP003726.1 | 100/96 | |
| ANC69 | Clostridium | AB161372.1 | 99/99 | |
| ANC71 | Bacillus | KP209387.1 | 100/97 | |
| ANC73 | Bacillus | CP009600.1 | 97/99 | |
| LB-1% CMC | CNC1 | Citrobacter | EU775609.1 | 100/92 |
| CNC4 | Lysinibacillus | KT254135.1 | 100/99 |
In this work, OTUs related to Bacillus and Lysinibacillus genera were found, similar to Montor-Antonio et al., 2014, although, in this case also OTUs related to Enterococcus, Clostridium and Citrobacter were also found. In deeper studies performed on Brazilian soils, a higher diversity, (43 genera) has been found (Pisa et al., 2011). However, in that research, DNA was isolated directly from soil samples. Due to the difference between libraries in the number of OTUs, the PCR products from LB-CMC and LB were cloned twice more, varying the insert concentration: however, the result remained the same (only two clones were obtained). The low number of OTUs found in the LB-CMC medium was possibly due to the brief incubation time (48 h), the substrate complexity (higher than starch), the enrichment adaptation phase (lag phase) and the number of sequences obtained from the library. Using microorganisms with cellulase genes or a cellulosome complex sometimes requires 10 days only to break down 50% of the cellulose (Yang et al., 2015). In this work, growth was stopped at 48 h. Yet, Citrobacter represents an important OTU in the degradation of cellulose with biotechnological relevance, as previous studies have found for H2 production in Citrobacter sp. in media enriched with cellobiose (Mangayil et al., 2011).
Our comparison is also limited by the methodology used, focussed on obtaining starch-and cellulose-related microbial diversity of soil with sugarcane crops, which can explain the low rates of diversity indexes and the specificity of the observed OTUs (Figure 6). However, the sequencing effort was adequate, since it was observed that at the end of the rarefaction curve it entered its plateau phase, indicating that it will be difficult to find more OTUs (Schloss and Handelsman, 2003). These results are expected because the cultivation conditions, rich in starch and cellulose, are not optimal for most microorganisms that inhabit the soil, more so for those specialized in their degradation (Montor-Antonio et al., 2014).
Phylogenetic classification of the library sequences obtained from the enrichment of soil samples
Ribotyping was used for the classification of the obtained OTUs that were assigned to a species level in some cases, or at least to a complex of really close species (Figures 1, 2, 3, 4 and 5). The most representative bacterial group was Clostridium with seventeen sequences, which form a new branch in some cases (Figure 4). Bacillus was represented by nine sequences, all in the B. anthracis, B. cereus and B. mycoides complex. In the Enterococcus genera, six sequences were found in the E. faecalis complex. In the Lysinibacillus genera, two sequences in the L. kistanensis, L. macroides and L. borotolerans complex were found, and one sequence was present in the Citrobacter genera phylogenetically related to C. werkmanii. Clostridium, Bacillus, Lysinibacillus and Enterococcus genera belong to the Firmicutes phylum; 97% of sequences obtained appertain to this phylum and only 3% to the Proteobacteria phylum. Some reports show that Firmicutes is the dominant phylum in sugarcane soil, mainly the Bacillus genus; however other research shows Proteobacteria (30%), Acidobacteria (23%), Bacteroides (12%) and Firmicutes (10%) as the principal reported phylum proportion (Pisa et al., 2011; Sharmin et al., 2013). Differences could exist due to type of soil and the methodology used.
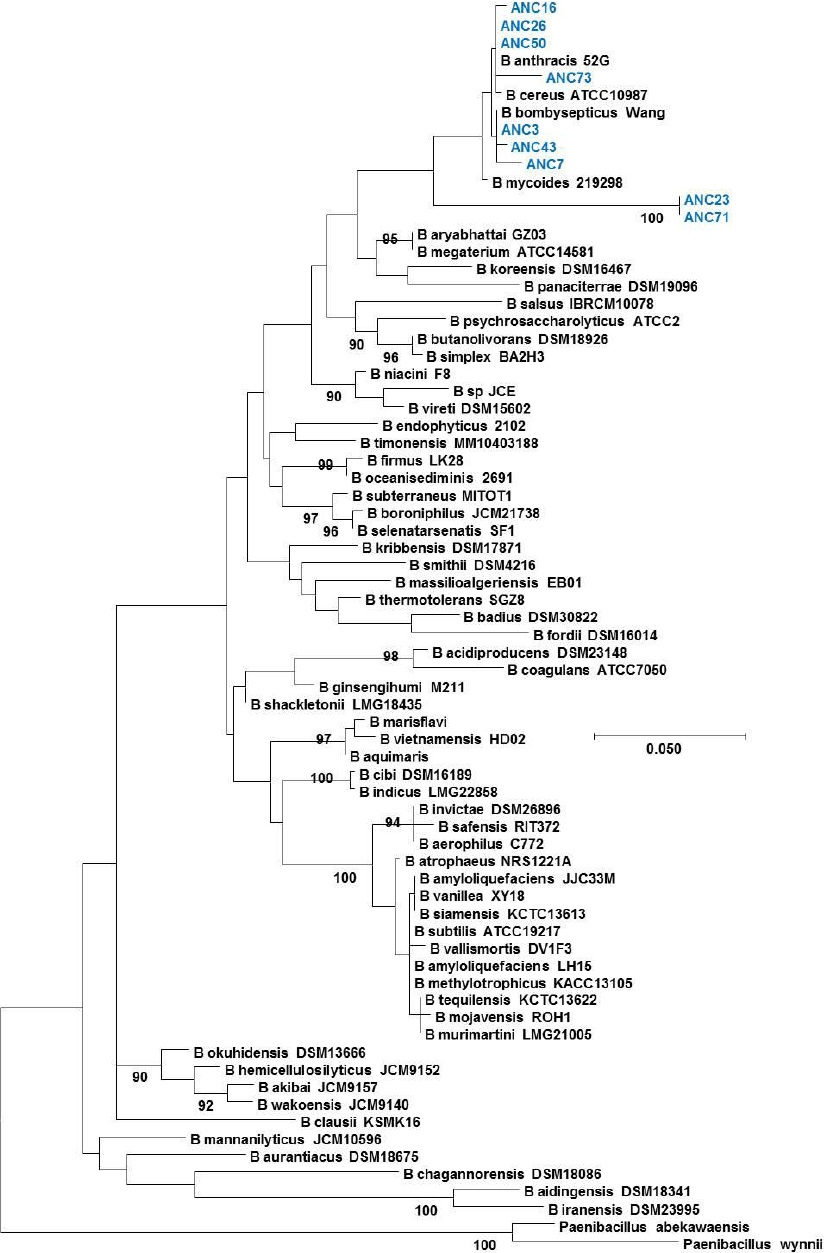
Figure 1 Phylogenetic tree of the Bacillus genus. Evolutionary inference was carried out following the maximum likelihood method, with the GTR + G (General Time Reversible Model plus Gamma distribution) model, and 500 bootstrap replicates. The Paenibacillus genus was used as the nearest external source.
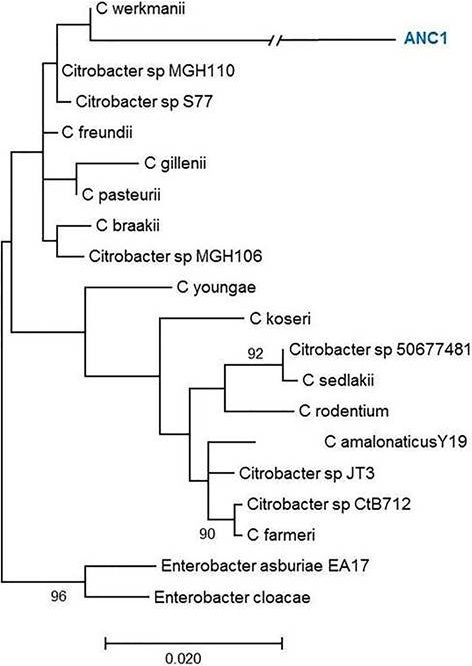
Figure 2 Phylogenetic tree of the Citrobacter genus. Evolutionary inference was carried out following the maximum likelihood method, with the GTR + G (General Time Reversible Model plus Gamma distribution) model, and 500 bootstrap replicates. The Enterobacter genus was used as the nearest external source.
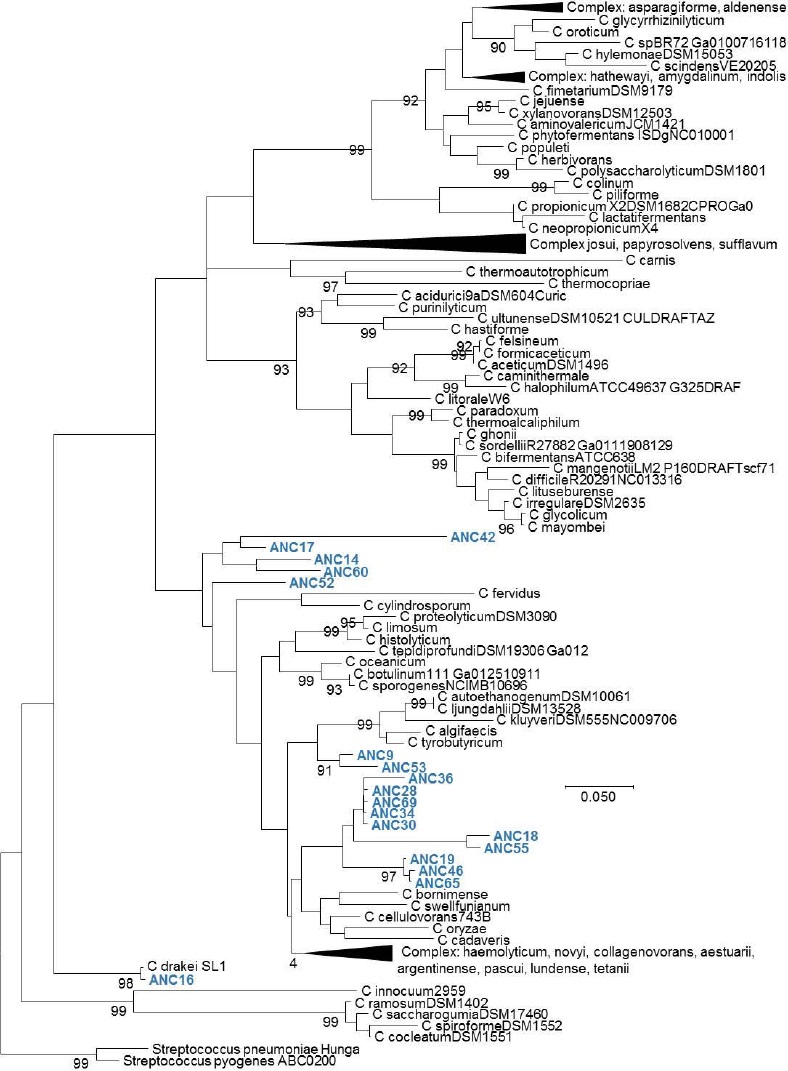
Figure 3 Phylogenetic tree of the Clostridium genus. Evolutionary inference was carried out following the maximum likelihood method, with the GTR + G (General Time Reversible Model plus Gamma distribution) model, and 500 bootstrap replicates. The Streptococcus genus was used as the nearest external source.
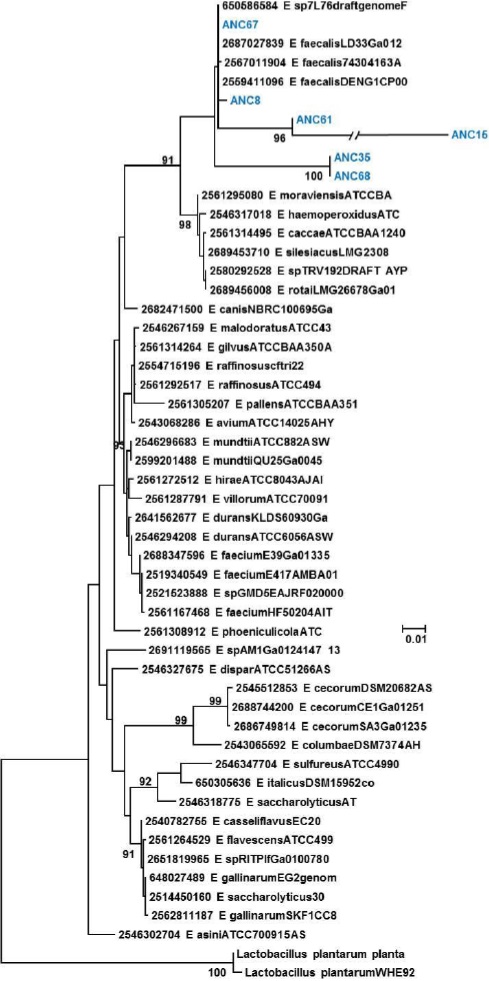
Figure 4 Phylogenetic tree of the Enterococcus genus. Evolutionary inference was carried out following the maximum likelihood method, with the GTR + G (General Time Reversible Model plus Gamma distribution) model, and 500 bootstrap replicates. The Lactobacillus genus was used as the nearest external source.
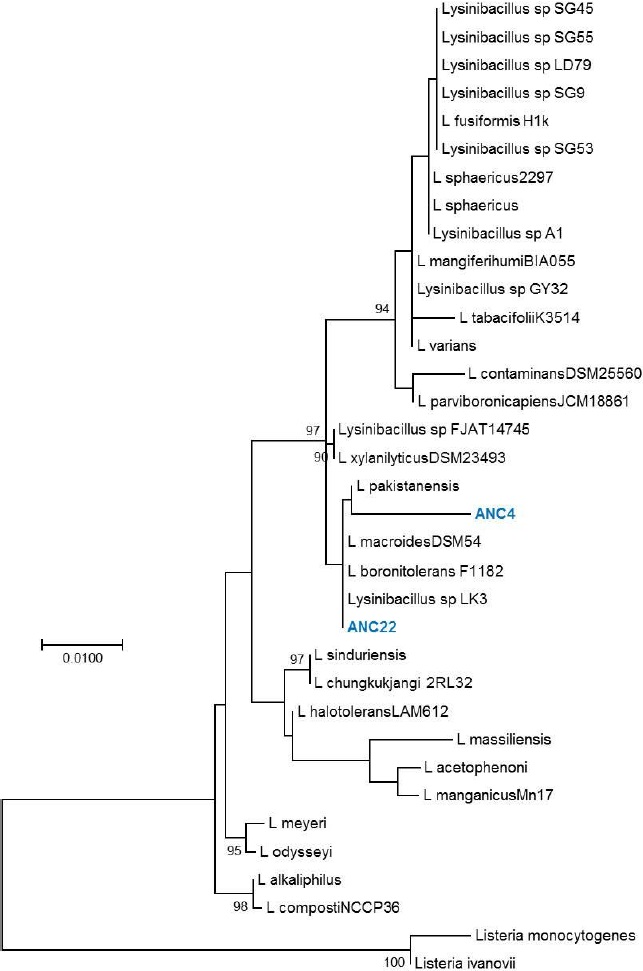
Figure 5 Phylogenetic tree of the Lysinibacillus genus. Evolutionary inference was carried out following the maximum likelihood method, with the GTR + G (General Time Reversible Model plus Gamma distribution) model, and 500 bootstrap replicates. The Listeria genus was used as the nearest external source.
The Firmicutes phylum including B. licheniformis, B. cereus, B. amyloliquefaciens and B. subtilis, produce amylolytic enzymes (α-amylase, α-glucosidase, amyloglucosidase) to break starch in the environment, to reduce Fe ions in anaerobiosis, and to use oxygen and nitrates as final electron acceptors (de Souza and Magalhães, 2010; Illmer and Schinner, 1992). Bacillus genus strains are related with phosphorus fixing, using several strategies, one of them being to produce acids to solubilize non-soluble phosphorus (Banik and Dey, 1982; Illmer and Schinner, 1992). Strains of B. licheniformis and B. amyloliquefaciens were found to produce mixtures of lactic, isovaleric, isobutyric, and acetic acids for that activity (Loredo-Osti, et al., 2004; Sivaramakrishnan et al., 2006). The Clostridium genus, for example C. acetobutylicum, C. butyricum and C. thermoamylolyticum, also includes microorganisms with α-amylase, amyloglucosidase genes able to degrade starch. Clostridium are obligatory anaerobic heterotrophs only capable of fixing N2 in the complete absence of oxygen (Kennedy et al., 2004; Kennedy and Tchan, 1992) and some Clostridium strains can reduce phosphate to phosphite in the soil (Almeida et al., 2011; Falkowski et al., 2008).
The Lysinibacillus genus is an endemic soil strain characterized by toxin production; genetically, it is related to the Bacillus genus and some strains, such as L. sphaericus show α-amylase activity (Kumar et al., 2012; Montor-Antonio et al., 2014; Tambekar et al., 2016). Enterococcus is part of the intestinal microbiota of animals and has been used as an indicator of fecal contamination in environmental samples; in fermented corn mass, a strain was identified with low α-amylase activity (Kumar et al., 2012; Mazzucotelli et al., 2013; Montor-Antonio et al., 2014; Tambekar et al., 2016). It should be noted that all these genera have strains which produce amylolytic enzymes. It is possible that the OTUs isolated in this work are related to starch degradation. The Citrobacter genus is important in the production of cellulases and H2 from lignocellulosic residues; in the soil, it is responsible for reducing nitrate to nitrite in the environment and phosphorus solubilization (Sprocati et al., 2014; Zhang et al., 2017). In this work, an OTU related to this species was only found in LB-CMC.
The bacteria genus here identified by OTUs belong to free-PGPB, demonstrated as having a symbiotic and enhanced relationship with rhizobacteria (Bashan et al., 1996; Reverbel-Leroy et al., 1996). Studies of bacteria such as Azotobacter, Azospirillum, Bacillus, and Klebsiella sp. are also used to inoculate a large area of arable land in the world with the aim of enhancing plant productivity (Lynch, 1983). In addition, phosphate solubilizing bacteria, such as the Bacillus and Paenibacillus species (formerly Bacillus), have been applied to soils to specifically enhance the phosphorus status of plants (Brown, 1974; Hayat et al., 2010). Therefore, isolating bacteria related to the Bacillus, Lysinibacillus and Clostridium genera can improve soil quality, as many belong to the free-PGPB group and are able to fix nitrogen, solubilize phosphate as well as degrading starch and cellulose, some of the most abundant polymers in the world.
Operational taxonomical units and the diversity indexes of the obtained sequences
The mothur program was run with the 35 obtained sequences, no chimera sequences were found, and 20 OTUs were observed at 97% of cutpoint (Figure 6). In general, in the rarefraction curve, a low diversity that agrees with the Chao, Shannon and Simpson indexes was observed (Figure 6).
Conclusions
With enriched metagenomic cultures, 35 OTUs were obtained, mainly from LB-starch; these are related to the Clostridium, Bacillus, Lysinibacillus and Enterococcus genera (Firmicutes). Also, an OTU related to Citrobacter was found, a genus important in cellulose degradation to H2 production. Soil tends to have a high organic material content, but according to the rarefaction curve it is not diverse, due to the use of complex substrates. Principally bacteria genera found in this study are related to free-living-PGPB. Some Bacillus and Citrobacter strains are highly involved with phosphorus solubilization and nitrogen fixing that correlates with the high content of phosphorus and nitrogen in the analyzed soil. Besides, the OTUs found are related to microorganisms able to break down starch and cellulose residues. The information acquired could be used by institutions or organizations related to sugarcane cultivation and commercialization to improve the soil quality via exogenous inoculation of the species here mentioned and for biotechnological applications in biofuel production.











 nueva página del texto (beta)
nueva página del texto (beta)



