Review articles
Genomic studies in epilepsy
-
Publication dates-
Jan-Mar , 2019
September 06, 2021
- Article in PDF
- Article in XML
- Automatic translation
- Send this article by e-mail
- Share this article +
Abstract
Epilepsy is a progressive and disabling disease if not diagnosed early; for this reason, it has been the subject of research, specially in cases with idiopathic etiology. Approximately between 1 and 2% of the world population have epilepsy. In Mexico the prevalence is from 10 to 20 patients per 1000 inhabitants. Lately, the scientific community has been trying to create, adapt, and use biomolecular tools to study its pathophysiology so that, hopefully, in a near future we are able to intervene in the natural history of this disease. The aim of this work is to cite evidence about some of the molecular biology techniques in order to support and encourage investment in neurogenomical research; as a necessary tool in the study of epilepsy.
Key words:
Epilepsy, Biomolecular Tools, Neurogenomics
Introduction
The International League Against Epilepsy (ILAE) defines an epileptic seizure as the occurrence of signs and/or symptoms due to an excessive synchronous or asynchronous abnormal neuronal activity, and epilepsy as a disease characterized by the long-term predisposition to generate epileptic seizures, as well as, by the neurobiological, cognitive, psychological, and social consequences of this condition1.
-
1ILAE official report:a practical clinical definition of epilepsyEpilepsia, 2014
In the world, there are >50 million people suffering from epilepsy, of which 80% live in developing countries with a prevalence of 7-14 per 1000 inhabitants, unlike the developed countries with a proportion of 4-10 per 1000 habitants2.
-
2OMS, Organización Mundial de la Salud,
In Mexico, a prevalence of 10-20 per 1000 habitants has been found; therefore, it can be estimated that there are approximately 1-2 million Mexicans affected3.
-
3Frecuencia en México,
Etiologically, epilepsy can be classified into the following groups: symptomatic or secondary (where there is a known cause, such as tumor, neuroinfection, and congenital brain malformation), idiopathic (when genetic factors are suspected, inherited, or de novo, etc.), and cryptogenic (type of epilepsy in which it cannot be associated to a certain cause)4. Around 20-30% of epilepsies are caused by acquired conditions and 70-80% are related to one or more genetic factors5.
-
4Epilpsy Society,
-
5Advancing epilepsy genetics in the genomic eraGenome Med, 2015
Epilepsy is considered a public health problem, due to its high morbidity and psychosocial repercussions (stigmatization or rejection) and economic (unemployment, pharmacological, and hospitalization expenses); therefore, it should be a reason for interest, investment, and research to understand the disease and provide the best care to this sector of the population. Considering the proportion of epilepsy related to genetic factors, 5 it is crucial to know the clinical, physiological, and genomic tools used in the diagnosis in this type of patients.
Since the publication in 1951 in JAMA by the epileptologist William G. Lennox, it was possible to confirm the importance of the genetic causes in some types of epilepsy, observed in their studies in twins6. Later, Watson and Crick (1953) propose the helical structure, antiparallel, and complementary to DNA7, researchers have used these principles for the development of molecular technology to understand and analyze the genetic material of all kinds of organisms, including humans, interest in the study of inheritance and genes, using genomics (a discipline that deals with the study of genomes, genes, and their functions, as well as related biotechnological techniques)8.
-
6The heredity of epilepsy as told by relatives and twinsJ Am Med Assoc, 1951
-
7Molecular structure of nucleic acids;A structure for deoxyribose nucleic acidNature, 1953
-
8Genomics and World Health:report of the Advisory Committee on Health Research, 2002
Advances in genomic technology are providing tools for the study of genetic factors that may be involved with different types of epilepsy. Some of the main types of studies used in epilepsy research are described below: full genome-wide association studies (GWAS), sequencing, next-generation sequencing (NGS), sequencing of whole genome (whole genome sequencing/[WGS]), complete exome sequencing (whole exome sequencing/[WES]), chromosomal microarrays (RNA and DNA microarrays) by comparative genomic hybridization (CGH) to detect copy number variations (CNVs), insertions and deletions, single-nucleotide polymorphisms (SNPs), or point mutations, (Fig. 1).
Thumbnail
Figure 1
Variations in the genome of a single base. Variants of a single nucleotide within a DNA sequence can be classified as SNP< 1% or as a point mutation <1% according to their frequency in the study population.
Variations in the genome of a single base. Variants of a single nucleotide within a DNA sequence can be classified as SNP< 1% or as a point mutation <1% according to their frequency in the study population.
Full Genome Association Studies (GWAS)
In genetic epidemiology, a complete genome association study (GWA) uses high-throughput technologies to analyze hundreds of thousands of SNPs (SNPs, generally referring to a single-base variant in the human genome) and relates them to measurable traits, as well as with various clinical conditions. These are studies designed to identify common genetic variants between two or more populations that contribute to a risk of disease9.
-
9Predictive rule inference for epistatic interaction detection in genome-wide association studiesBioinformatics, 2010
As an example, in 2014, the ILAE published a meta-analysis of 12 cohorts where they performed a complete genome association on 8696 patients with epilepsy and 26,157 controls. They found association of risk in the loci 2q24.3 (p=8.71×10−10) that involves the gene SCN1A and in the loci 4p15.1 (p=5 44×10−9) that involves the PCDH7 gene in patients with focal and generalized epilepsy. For patients with generalized epilepsy at the 2p16.1 loci (p=9.99×10−9), which implicate the VRK2 or FANCL genes, they could not determine an SNP with statistical significance related to focal epilepsy10. Feenstra et al.11 studied through GWA children with febrile seizures as an adverse effect after the administration of the triple viral vaccine (rubella, measles, and mumps), children who did not have febrile seizures after the vaccine and finally children without a history of febrile seizures as controls. They found two risk loci related to febrile seizures after vaccination rs273259 (p=5.9×10−12 and p=1.2×10−9) involving the gene IFI44L and rs1318653 (p=9.6×10−11 and p=1.6×10−9) that involves the CD46 gene, with p values against the controls and against children who did not have febrile seizures after the vaccine, respectively. On the other hand, they found four risk loci for febrile seizures, in general, two were in known genes related to epilepsy (SCN1A and SCN2A).
-
10Genetic determinants of common epilepsies:a meta-analysis of genome-wide association studiesLancet Neurol, 2014
-
11Common variants associated with general and MMR vaccine-related febrile seizuresNat Genet, 2014
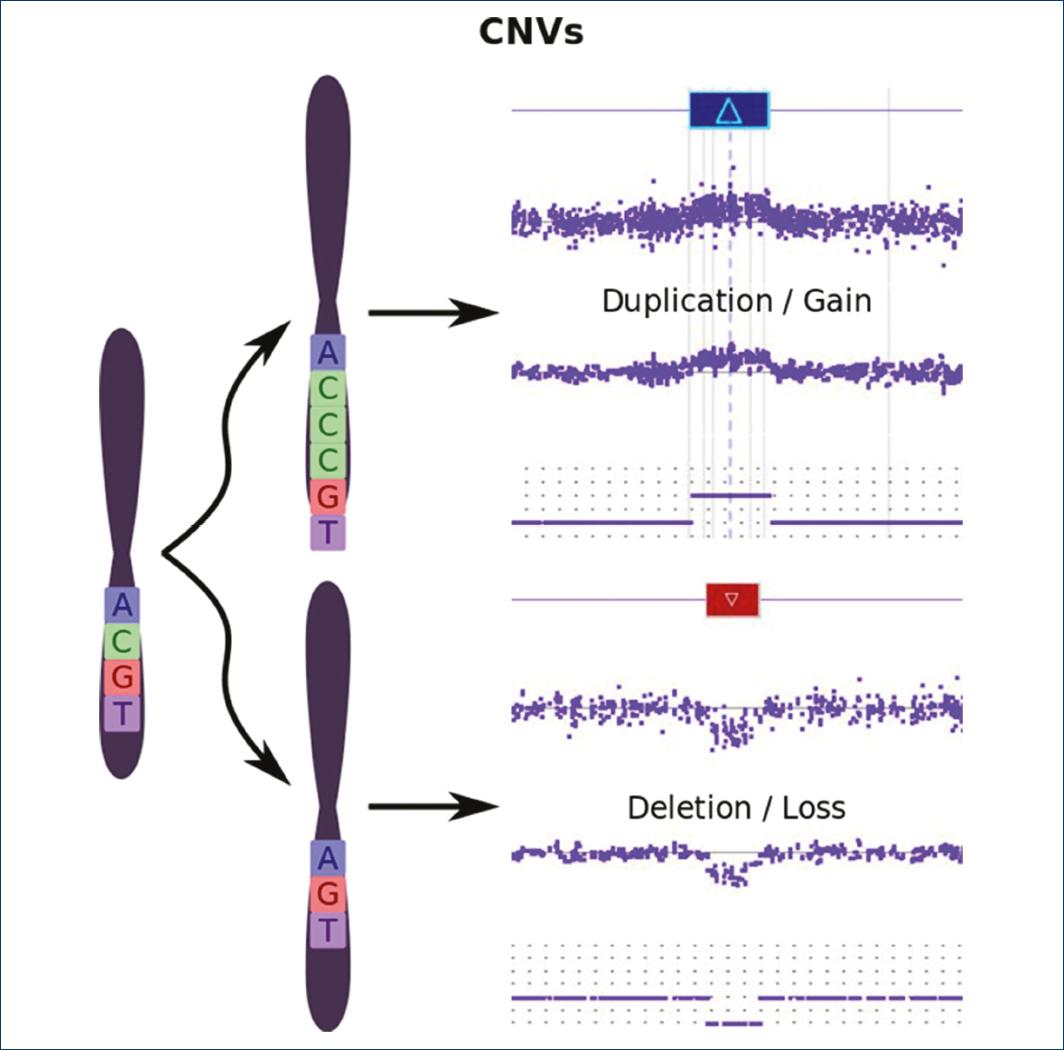
Copy Number Variations (CNVs)
CNVs are defined as a DNA segment equal to or >1 kb whose number of copies is variable (duplicated or deleted) when compared to a reference genome (Fig. 2).
Thumbnail
Figure 2
Copy number variations (CNVs). Variation in the number of copies, by loss (deletion) or gain (duplication) of a DNA segment greater than one kilobase with respect to a reference genome.
Copy number variations (CNVs). Variation in the number of copies, by loss (deletion) or gain (duplication) of a DNA segment greater than one kilobase with respect to a reference genome.
CNVs are an important source of normal genetic variation (in a frequency >1%), but some may participate as risk factors or causes of disease5,12. CNVs can be detected with DNA microarrays by means of CGH, array CGH, (Fig. 3)
-
5Advancing epilepsy genetics in the genomic eraGenome Med, 2015
-
12Mechanisms of change in gene copy numberNat Rev Genet, 2009
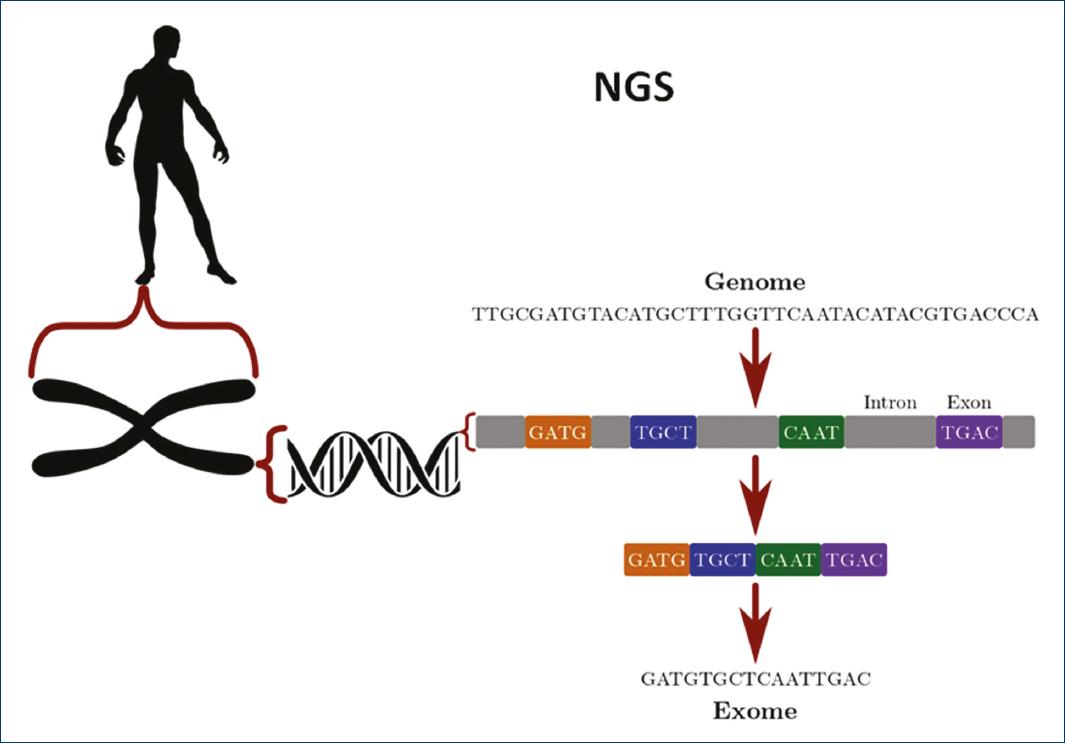
Thumbnail
Figure 3
Next-generation sequencing (NGS) determines the order of the nucleotides of a specific sequence. It can be used to determine only the coding part of a sequence (exome) or the whole sequence (genome).
Next-generation sequencing (NGS) determines the order of the nucleotides of a specific sequence. It can be used to determine only the coding part of a sequence (exome) or the whole sequence (genome).
Some rare CNVs (frequency <1%) involve genes from known diseases and may be related to 5-10% in cases of childhood epilepsy13,14. Helbig et al. reported the role of CNVs in patients with epilepsy, finding 88 rare NVs in 71 patients (31.8%) >100 kb related to the disease15.
-
13Copy number variation plays an important role in clinical epilepsyAnn Neurol, 2014
-
14Rare copy number variants are an important cause of epileptic encephalopathiesAnn Neurol, 2011
-
15Structural genomic variation in childhood epilepsies with complex phenotypesEur J Hum Genet, 2014
In general, research in generalized and focal epilepsy has identified recurrent microdeletions in up to 3% of patients with idiopathic generalized epilepsy and 1% focal epilepsy. The microdeletions in the chromosomal regions 15q13.3 and 16p13.11 are the most frequently identified variants16,17.
-
16Genome-wide copy number variation in epilepsy:novel susceptibility loci in idiopathic generalized and focal epilepsiesPLoS Genet, 2010
-
17Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromesAm J Hum Genet, 2010
Next Generation Sequencing (NGS)
DNA sequencing refers to the determination of the order of the nucleotides of a given sequence, from some base pairs (bp) to the sequence of complete genomes. The NGS, also called mass sequencing in parallel, means that millions of small DNA fragments (around 100 bp) can be sequenced at the same time18.
-
18Secuenciación de genoma completo:un salto cualitativo en los estudios genéticosRev Neurol, 2012
At present, two types of sequencing are performed for the study of epilepsy: complete genome sequencing and complete exome sequencing (Fig. 3).
Complete Genome Sequencing (WGS)
It refers to the determination of the order of the nucleotides of the whole genome (both the coding and non-coding sequences) which covers around 3000 million bp19.
-
19Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosisHum Mol Genet, 2014
Complete Exome Sequencing (WES)
This technique allows exploring 180,000 exons or coding regions (more or less 30 million bp), which corresponds to approximately 1% of the human genome20, it is estimated that 85% of the variations related to hereditary diseases are found in the exome18.
-
20Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosisHum Mol Genet, 2014
-
18Secuenciación de genoma completo:un salto cualitativo en los estudios genéticosRev Neurol, 2012
Helbig et al. evaluated the performance of exome sequencing as a diagnostic method in patients with epilepsy, finding 38.2% positive results compared to controls with p=0.004 value, concluding that this technique is a useful diagnostic tool, especially in severe epilepsy of early onset21.
-
21Targeted capture and massively parallel sequencing of the human exomeJ Investig Med, 2010
In the past 10 years, the advancement of complete genome sequencing or exome techniques has allowed the identification of new genes and genetic variants involved in family epilepsies, severe epilepsies, and epileptic encephalopathies, which has had an important impact in the diagnosis of this disease. The current rate in the diagnosis of epilepsy by NGS ranges from 20% to 30% and specifically with WES is approximately 25%22.
-
22Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsyGenet Med, 2016
Candidate Genes Related to Epilepsy
Some of the major genes involved in generalized epilepsy are described below:
SCN1A codes for the alpha-1 subunit of the voltage-dependent sodium channel. The transmembrane alpha subunit forms the central pore of the channel. This ion channel is critical for the generation and propagation of action potentials. The channel responds to the voltage difference across the cell membrane to create a pore that allows sodium ions to pass through the membrane. The influx of sodium creates an action potential, which is critical for signaling within the brain. Mutations of loss of function cause a reduction of sodium currents and alteration of the signaling of the hippocampal GABAergic interneurons. Allelic variants of this gene are associated with generalized epilepsy with febrile seizures and epileptic encephalopathy. In 70-90% of cases, Dravet syndrome is caused by a de novo mutation in SCN1A, which often leads to a non-functional protein23,24.
-
23The contribution of next generation sequencing to epilepsy geneticsExpert Rev Mol Diagn, 2015
-
24Beyond the Ion Channel,
SCN2A encodes the alpha-II subunit of the voltage-dependent sodium channel and is found in the initial segment of the axon, nonsense mutations are observed in patients with epileptic encephalopathies where their expression is reduced on the cell surface, resulting in a net loss of function. This mutation is related to four different phenotypes such as benign neonatal and infantile epilepsy, autism and intellectual disability, infantile spasms, and early-onset epileptic encephalopathies including Ohtahara syndrome and severe neonatal epilepsy. All phenotypes within the SCN2A spectrum include cognitive disturbances, seizures, and movement disorders23,24.
-
23The contribution of next generation sequencing to epilepsy geneticsExpert Rev Mol Diagn, 2015
-
24Beyond the Ion Channel,
CACNA1A codes for the alpha-1 subunit of voltage-dependent calcium channels and mediates the entry of calcium ions into excitable cells; it is also included calcium-dependent processes including muscle contraction, hormone release, and neurotransmitter release. Mutations in this gene are related to episodic ataxias, spinocerebellar degeneration, and familial hemiplegic migraine, generalized epilepsies such as absences or Dravet syndrome, and tonic paroxysms23,24.
-
23The contribution of next generation sequencing to epilepsy geneticsExpert Rev Mol Diagn, 2015
-
24Beyond the Ion Channel,
Regarding focal epilepsy, the main candidate genes are described below:
GRIN2A encodes the alpha-2 subunit of the glutamate receptor N-methyl-D-aspartate, it is involved in long-term potentiation, an activity-dependent increase in the efficiency of synaptic transmission; the interruption of this gene is associated with the disorder of focal rolandic epilepsy, atypical benign partial epilepsy, Landau-Kleffner syndrome, and some learning disorders23,24.
-
23The contribution of next generation sequencing to epilepsy geneticsExpert Rev Mol Diagn, 2015
-
24Beyond the Ion Channel,
DEPDC5 codes for a member of the IML1 family of proteins involved in G-protein signaling pathways (mTORC1) and regulates cell growth by detecting nutrient availability; inhibition of mTOR can cause cortical dysplasia at variable sites. Mutations in this gene have been related to focal epilepsy of variable foci, nocturnal frontal lobe dominant epilepsy, and temporal mesial lobe family epilepsy23,24.
-
23The contribution of next generation sequencing to epilepsy geneticsExpert Rev Mol Diagn, 2015
-
24Beyond the Ion Channel,
LGI1 gene codes for a member of the superfamily of proteins rich in leucine (glioma rich in inactivated leucine), can regulate the activity of voltage-dependent potassium channels, and is involved in the regulation of neuronal growth and cell survival. This gene is rearranged as the result of translocations in glioblastoma cell lines. Mutations in this gene are related to lateral temporal epilepsy23,24.
-
23The contribution of next generation sequencing to epilepsy geneticsExpert Rev Mol Diagn, 2015
-
24Beyond the Ion Channel,
The discovery of mutations in specific genes (encoders for ion channels expressed in brain neurons, neurotransmitter receptors, or molecules with assumed functions in intercellular communication) has allowed to corroborate the suspicions that the physiopathological bases of this disease seem to be related with alterations in the electrical type processes, especially those that cause alterations in the stability of the membranes25,26.
-
25Gene Cards, 2016
-
26Genetics advances in autosomal dominant focal epilepsies:focus on DEPDC5Prog Brain Res, 2014
The table summarizes some of the candidate genes related to epilepsy, discovered by sequencing, association studies, DNA microarrays, etc. (Supplementary Table 1).
One of the main goals in the molecular research of epilepsy is to provide personalized treatment, and some data are beginning to emerge that this may be possible, in 2014, the abnormal gain of the function of the KCNQ1 gene that codes for member 1 of the Q subfamily of potassium channels dependent on filtration and reverts with quinidine27. On the other hand, personalized therapy with memantine or topiramate was also proposed in two patients with early-onset epileptic encephalopathy with mutation in the GRIN2A gene28.
-
27Benign mesial temporal lobe epilepsyNat Rev Neurol, 2011
-
28KCNT1 gain of function in 2 epilepsy phenotypes is reversed by quinidineAnn Neurol, 2014
It is important to take into account the genetic factors related to the disease when deciding the treatment of the patient, especially if the treatment is a surgical procedure. Skjei et al. published a series of cases in which they describe the clinical and histopathological characteristics in six patients with refractory epilepsy and mutations in the SCN1A gene undergoing focal cortical resection. In all cases, patients were refractory to the surgical procedure; it was observed mild diffuse malformations of cortical development in four of six patients concluding that cortical resection may not be effective in patients with this mutation and with the neuropathological changes mentioned29.
-
29GRIN2A mutation and early-onset epileptic encephalopathy:personalized therapy with memantineAnn Clin Transl Neurol, 2014
New approaches for the treatment of epilepsy are under development, experimental research based on viral vectors, genetic opto tools involving the use of light at wavelengths of 280-570 nm, to control the activity of ion channels in rhodopsin and halorhodopsin in hippocampal neurons, dentate gyrus, and cerebellum, which activate or inhibit a neuron and even several conglomerates of neuronal networks that allow a control of neuronal electrical activity and cell graft techniques in animal models; all of them are new techniques used for a future to prevent the disease or to provide the best treatment to this type of patients30,31.
-
30Clinical and histopathological outcomes in patients with SCN1A mutations undergoing surgery for epilepsyJ Neurosurg Pediatr, 2015
-
31Recent advances in epilepsyJ Neurol, 2014
Conclusion
Epilepsy is considered a disease of complex inheritance, the main difficulties associated with the study of complex diseases are incomplete penetrance, genetic heterogeneity, and polygenic (or multifactorial) inheritance32. Therefore, it is not yet clear what is the role of inheritance and other genetic factors in epileptogenesis. There is currently a project called Phenotype/Epilepsy Genotype EPGP: the Epilepsy Phenome/Genome Project; it is a large-scale project involving 27 centers in the United States, Australia, Argentina and Canada with the aims of analyzing the detailed phenotype of patients, determining the genotype and discovering new genes. Genetics and genomics in epilepsy is an open field of research that has had a great break in the last 15 years, which has solved many of the cases that were previously classified as of unknown cause, however, there is still a long way to go.
-
32Modelos experimentales en optogenética y su aplicación en enfermedades neurodegenerativas motorasRev Med Inv, 2015
References
-
1Fisher RS, Acevedo C, Arzimanoglou A, et al. ILAE official report:a practical clinical definition of epilepsy. Epilepsia. 2014;55:475-82. Links
-
2OMS, Organización Mundial de la Salud. Available from:http://www.who.int/mediacentre/factsheets/fs999/en/. [Last accessed on 2016 Jul 17]. Links
-
3Programa Prioritario de Epilepsia. Frecuencia en México. Available from:http://www.epilepsiamexico.gob.mx/info-pacientes/frecuencia.htm. [Last accessed on 2016 Jul 17]. Links
-
4Epilpsy Society. Available from:https://www.epilepsysociety.org.uk/causes-epilepsy#.V8MuCSYoDtQ. [Last accessed on 2016 Jul 17]. Links
-
5Myers CT, Mefford HC. Advancing epilepsy genetics in the genomic era. Genome Med. 2015;7:91. Links
-
6Lennox WG. The heredity of epilepsy as told by relatives and twins. J Am Med Assoc. 1951;146:529-36. Links
-
7Watson JD, Crick FH. Molecular structure of nucleic acids;A structure for deoxyribose nucleic acid. Nature. 1953;171:737-8. Links
-
8Genomics and World Health:report of the Advisory Committee on Health Research. Geneva:WHO;2002. Available from:http://www.who.int/genomics/geneticsVSgenomics/en/. [Last accessed on 2016 Jul 17]. Links
-
9Wan X, Yang C, Yang Q, et al. Predictive rule inference for epistatic interaction detection in genome-wide association studies. Bioinformatics. 2010;26:30-7. Links
-
10International League Against Epilepsy Consortium on Complex Epilepsies. Electronic address:epilepsy-austin@unimelb.edu.au. Genetic determinants of common epilepsies:a meta-analysis of genome-wide association studies. Lancet Neurol. 2014;13:893-903. Links
-
11Feenstra B, Pasternak B, Geller F, et al. Common variants associated with general and MMR vaccine-related febrile seizures. Nat Genet. 2014;46:1274-82. Links
-
12Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551-64. Links
-
13Olson H, Shen Y, Avallone J, et al. Copy number variation plays an important role in clinical epilepsy. Ann Neurol. 2014;75:943-58. Links
-
14Mefford HC, Yendle SC, Hsu C, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Ann Neurol. 2011;70:974-85. Links
-
15Helbig I, Swinkels ME, Aten E, et al. Structural genomic variation in childhood epilepsies with complex phenotypes. Eur J Hum Genet. 2014;22:896-901. Links
-
16Mefford HC, Muhle H, Ostertag P, et al. Genome-wide copy number variation in epilepsy:novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. Links
-
17Heinzen EL, Radtke RA, Urban TJ, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet. 2010;86:707-18. Links
-
18Jiménez-Escrig A, Gobernado I, Sánchez-Herranz A. Secuenciación de genoma completo:un salto cualitativo en los estudios genéticos. Rev Neurol. 2012;54:692-8. Links
-
19Martin HC, Kim GE, Pagnamenta AT, et al. Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis. Hum Mol Genet. 2014;23:3200-11. Links
-
20Kumar A, Turner E, Shendure J. Targeted capture and massively parallel sequencing of the human exome. J Investig Med. 2010;58:123. Links
-
21Helbig KL, Farwell Hagman KD, Shinde DN, et al. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med. 2016;18:898-905. Links
-
22Møller RS, Dahl HA, Helbig I. The contribution of next generation sequencing to epilepsy genetics. Expert Rev Mol Diagn. 2015;15:1531-8. Links
-
23The ILAE Genetics Commission Blog. Beyond the Ion Channel. Available from:http://www.epilepsygenetics.net/the-epilepsiome/scn1a-this-is-what-you-need-to-know/. [Last accessed on 2016 Mar 17]. Links
-
24Gene Cards. Available from:http://www.genecards.org/, 2016. [Last accessed on 2016 Mar 04]. Links
-
25Baulac S. Genetics advances in autosomal dominant focal epilepsies:focus on DEPDC5. Prog Brain Res. 2014;213:123-39. Links
-
26Labate A, Gambardella A, Andermann E, et al. Benign mesial temporal lobe epilepsy. Nat Rev Neurol. 2011;7:237-40. Links
-
27Milligan CJ, Li M, Gazina EV, et al. KCNT1 gain of function in 2 epilepsy phenotypes is reversed by quinidine. Ann Neurol. 2014;75:581-90. Links
-
28Pierson TM, Yuan H, Marsh ED, et al. GRIN2A mutation and early-onset epileptic encephalopathy:personalized therapy with memantine. Ann Clin Transl Neurol. 2014;1:190-8. Links
-
29Skjei KL, Church EW, Harding BN, et al. Clinical and histopathological outcomes in patients with SCN1A mutations undergoing surgery for epilepsy. J Neurosurg Pediatr. 2015;16:668-74. Links
-
30Kovac S, Walker MC. Recent advances in epilepsy. J Neurol. 2014;261:837-41. Links
-
31Ortiz-Vilchis CM. Modelos experimentales en optogenética y su aplicación en enfermedades neurodegenerativas motoras. Rev Med Inv. 2015;3:107-68. Links
-
32Lopes-Cendes I, Ribeiro PA. Aspectos genéticos de las epilepsias:una visión actualizada. Rev Méd Clín Las Condes. 2013;24:909-14. Links
Supplementary material references
-
1de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921-9. Links
-
2Dimassi S, Labalme A, Ville D, et al. Whole-exome sequencing improves the diagnosis yield in sporadic infantile spasm syndrome. Clin Genet. 2016;89:198-204. Links
-
3Epi4K Consortium, Epilepsy Phenome/Genome Project, Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217-21. Links
-
4Michaud JL, Lachance M, Hamdan FF, et al. The genetic landscape of infantile spasms. Hum Mol Genet. 2014;23:4846-58. Links
-
5Møller RS, Dahl HA, Helbig I. The contribution of next generation sequencing to epilepsy genetics. Expert Rev Mol Diagn 2015;15:1531-8. Links
-
6Timal S, Hoischen A, Lehle L, et al. Gene identification in the congenital disorders of glycosylation Type I by whole-exome sequencing. Hum Mol Genet. 2012;21:4151-61. Links
-
7Harvey K, Duguid IC, Alldred MJ, et al. The GDP-GTP exchange factor collybistin:an essential determinant of neuronal gephyrin clustering. J Neurosci. 2004;24:5816-26. Links
-
8Lesca G, Till M, Labalme A, Vallee D, et al. De novo xq11.11 microdeletion including ARHGEF9 in a boy with mental retardation, epilepsy, macrosomia, and dysmorphic features. Am J Med Genet A. 2011;155A:1706-11. Links
-
9Lesca G, Depienne C. Epilepsy genetics:the ongoing revolution. Rev Neurol. 2015;171:539-57. Links
-
10Shimojima K, Sugawara M, Shichiji M, et al. Loss-of-function mutation of collybistin is responsible for X-linked mental retardation associated with epilepsy. J Hum Genet. 2011;56:561-5. Links
-
11Bruyere H, Lewis S, Wood S, MacLeod PJ, Langlois S. Confirmation of linkage in X-linked infantile spasms (West syndrome) and refinement of the disease locus to xp21.3-xp22.1. Clin Genet. 1999;55:173-81. Links
-
12Claes S, Devriendt K, Lagae L, et al. The X-linked infantile spasms syndrome (MIM 308350) maps to xp11.4-xpter in two pedigrees. Ann Neurol. 1997;42:360-4. Links
-
13Feinberg AP, Leahy WR. Infantile spasms:case report of sex-linked inheritance. Dev Med Child Neurol. 1977;19:524-6. Links
-
14Fullston T, Brueton L, Willis T, et al. Ohtahara syndrome in a family with an ARX protein truncation mutation (c.81C>G/p.Y27X). Eur J Hum Genet. 2010;18:157-62. Links
-
15Giordano L, Sartori S, Russo S, et al. Familial ohtahara syndrome due to a novel ARX gene mutation. Am J Med Genet A. 2010;152A:3133-7. Links
-
16Kato M, Das S, Petras K, et al. Mutations of ARX are associated with striking pleiotropy and consistent genotype-phenotype correlation. Hum Mutat. 2004;23:147-59. Links
-
17Kato M, Saitoh S, Kamei A, et al. A longer polyalanine expansion mutation in the ARX gene causes early infantile epileptic encephalopathy with suppression-burst pattern (Ohtahara syndrome). Am J Hum Genet. 2007;81:361-6. Links
-
18Poeta L, Fusco F, Drongitis D, et al. A regulatory path associated with X-linked intellectual disability and epilepsy links KDM5C to the polyalanine expansions in ARX. Am J Hum Genet. 2013;92:114-25. Links
-
19Proud VK, Levine C, Carpenter NJ. New X-linked syndrome with seizures, acquired micrencephaly, and agenesis of the corpus callosum. Am J Med Genet. 1992;43:458-66. Links
-
20Strømme P, Mangelsdorf ME, Shaw MA, et al. Mutations in the human ortholog of aristaless cause X-linked mental retardation and epilepsy. Nat Genet. 2002;30:441-5. Links
-
21Strømme P, Sundet K, Mørk C, et al. X linked mental retardation and infantile spasms in a family:new clinical data and linkage to xp11.4-xp22.11. J Med Genet. 1999;36:374-8. Links
-
22Turner G, Partington M, Kerr B, Mangelsdorf M, Gecz J. Variable expression of mental retardation, autism, seizures, and dystonic hand movements in two families with an identical ARX gene mutation. Am J Med Genet. 2002;112:405-11. Links
-
23Chan YC, Burgunder JM, Wilder-Smith E, et al. Electroencephalographic changes and seizures in familial hemiplegic migraine patients with the CACNA1A gene S218L mutation. J Clin Neurosci. 2008;15:891-4. Links
-
24Chioza B, Wilkie H, Nashef L, et al. Association between the alpha(1a) calcium channel gene CACNA1A and idiopathic generalized epilepsy. Neurology. 2001;56:1245-6. Links
-
25Holtmann M, Opp J, Tokarzewski M, Korn-Merker E. Human epilepsy, episodic ataxia Type 2, and migraine. Lancet. 2002;359:170-1. Links
-
26Jouvenceau A, Eunson LH, Spauschus A, et al. Human epilepsy associated with dysfunction of the brain P/Q-type calcium channel. Lancet. 2001;358:801-7. Links
-
27Kors EE, Melberg A, Vanmolkot KR, et al. Childhood epilepsy, familial hemiplegic migraine, cerebellar ataxia, and a new CACNA1A mutation. Neurology. 2004;63:1136-7. Links
-
28Chen Y, Lu J, Pan H, et al. Association between genetic variation of CACNA1H and childhood absence epilepsy. Ann Neurol. 2003;54:239-43. Links
-
29Heron SE, Khosravani H, Varela D, et al. Extended spectrum of idiopathic generalized epilepsies associated with CACNA1H functional variants. Ann Neurol. 2007;62:560-8. Links
-
30Khosravani H, Altier C, Simms B, et al. Gating effects of mutations in the cav3.2 T-type calcium channel associated with childhood absence epilepsy. J Biol Chem. 2004;279:9681-4. Links
-
31Khosravani H, Bladen C, Parker DB, et al. Effects of cav3.2 channel mutations linked to idiopathic generalized epilepsy. Ann Neurol. 2005;57:745-9. Links
-
32Vitko I, Chen Y, Arias JM, et al. Functional characterization and neuronal modeling of the effects of childhood absence epilepsy variants of CACNA1H, a T-type calcium channel. J Neurosci. 2005;25:4844-55. Links
-
33Mefford HC, Yendle SC, Hsu C, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Ann Neurol. 2011;70:974-85. Links
-
34Vergult S, Dheedene A, Meurs A, et al. Genomic aberrations of the CACNA2D1 gene in three patients with epilepsy and intellectual disability. Eur J Hum Genet. 2015;23:628-32. Links
-
35Archer HL, Evans J, Edwards S, et al. CDKL5 mutations cause infantile spasms, early onset seizures, and severe mental retardation in female patients. J Med Genet. 2006;43:729-34. Links
-
36Elia M, Falco M, Ferri R, et al. CDKL5 mutations in boys with severe encephalopathy and early-onset intractable epilepsy. Neurology. 2008;71:997-9. Links
-
37Bartnik M, Derwińska K, Gos M, et al. Early-onset seizures due to mosaic exonic deletions of CDKL5 in a male and two females. Genet Med. 2011;13:447-52. Links
-
38Erez A, Patel AJ, Wang X, et al. Alu-specific microhomology-mediated deletions in CDKL5 in females with early-onset seizure disorder. Neurogenetics. 2009;10:363-9. Links
-
39Kalscheuer VM, Tao J, Donnelly A, et al. Disruption of the serine/threonine kinase 9 gene causes severe X-linked infantile spasms and mental retardation. Am J Hum Genet. 2003;72:1401-11. Links
-
40Nectoux J, Heron D, Tallot M, Chelly J, Bienvenu T. Maternal origin of a novel C-terminal truncation mutation in CDKL5 causing a severe atypical form of rett syndrome. Clin Genet. 2006;70:29-33. Links
-
41Nemos C, Lambert L, Giuliano F, et al. Mutational spectrum of CDKL5 in early-onset encephalopathies:a study of a large collection of French patients and review of the literature. Clin Genet. 2009;76:357-71. Links
-
42Rademacher N, Hambrock M, Fischer U, et al. Identification of a novel CDKL5 exon and pathogenic mutations in patients with severe mental retardation, early-onset seizures and rett-like features. Neurogenetics. 2011;12:165-7. Links
-
43Rosas-Vargas H, Bahi-Buisson N, Philippe C, et al. Impairment of CDKL5 nuclear localisation as a cause for severe infantile encephalopathy. J Med Genet. 2008;45:172-8. Links
-
44Russo S, Marchi M, Cogliati F, et al. Novel mutations in the CDKL5 gene, predicted effects and associated phenotypes. Neurogenetics. 2009;10:241-50. Links
-
45Saletti V, Canafoglia L, Cambiaso P, et al. A CDKL5 mutated child with precocious puberty. Am J Med Genet A. 2009;149A:1046-51. Links
-
46Scala E, Ariani F, Mari F, et al. CDKL5/STK9 is mutated in rett syndrome variant with infantile spasms. J Med Genet. 2005;42:103-7. Links
-
47Tao J, Van Esch H, Hagedorn-Greiwe M, et al. Mutations in the X-linked cyclin-dependent kinase-like 5 (CDKL5/STK9) gene are associated with severe neurodevelopmental retardation. Am J Hum Genet. 2004;75:1149-54. Links
-
48Van Esch H, Jansen A, Bauters M, Froyen G, Fryns JP. Encephalopathy and bilateral cataract in a boy with an interstitial deletion of xp22 comprising the CDKL5 and NHS genes. Am J Med Genet A. 2007;143:364-9. Links
-
49Weaving LS, Christodoulou J, Williamson SL, et al. Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am J Hum Genet. 2004;75:1079-93. Links
-
50Carvill GL, Heavin SB, Yendle SC, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45:825-30. Links
-
51Chénier S, Yoon G, Argiropoulos B, et al. CHD2 haploinsufficiency is associated with developmental delay, intellectual disability, epilepsy and neurobehavioural problems. J Neurodev Disord. 2014;6:9. Links
-
52Galizia EC, Myers CT, Leu C, et al. CHD2 variants are a risk factor for photosensitivity in epilepsy. Brain. 2015;138:1198-207. Links
-
53Liu JC, Ferreira CG, Yusufzai T. Human CHD2 is a chromatin assembly ATPase regulated by its chromo-and DNA-binding domains. J Biol Chem. 2015;290:25-34. Links
-
54Lund C, Brodtkorb E, Øye AM, Røsby O, Selmer KK. CHD2 mutations in lennox-gastaut syndrome. Epilepsy Behav. 2014;33:18-21. Links
-
55Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability:an exome sequencing study. Lancet. 2012;380:1674-82. Links
-
56Suls A, Jaehn JA, Kecskés A, et al. De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with dravet syndrome. Am J Hum Genet. 2013;93:967-75. Links
-
57Thomas RH, Zhang LM, Carvill GL, et al. CHD2 myoclonic encephalopathy is frequently associated with self-induced seizures. Neurology. 2015;84:951-8. Links
-
58Trivisano M, Striano P, Sartorelli J, et al. CHD2 mutations are a rare cause of generalized epilepsy with myoclonic-atonic seizures. Epilepsy Behav. 2015;51:53-6. Links
-
59Dibbens LM, Mullen S, Helbig I, et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy:precedent for disorders with complex inheritance. Hum Mol Genet. 2009;18:3626-31. Links
-
60Helbig I, Mefford HC, Sharp AJ, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160-2. Links
-
61Peñagarikano O, Abrahams BS, Herman EI, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235-46. Links
-
62Strauss KA, Puffenberger EG, Huentelman MJ, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370-7. Links
-
63Alakurtti K, Weber E, Rinne R, et al. Loss of lysosomal association of cystatin B proteins representing progressive myoclonus epilepsy, EPM1, mutations. Eur J Hum Genet. 2005;13:208-15. Links
-
64Bespalova IN, Adkins S, Pranzatelli M, Burmeister M. Novel cystatin B mutation and diagnostic PCR assay in an unverricht-lundborg progressive myoclonus epilepsy patient. Am J Med Genet. 1997;74:467-71. Links
-
65de Haan GJ, Halley DJ, Doelman JC, et al. Univerricht-lundborg (sic) disease:underdiagnosed in the Netherlands. Epilepsia. 2004;45:1061-3. Links
-
66Di Giaimo R, Riccio M, Santi S, et al. New insights into the molecular basis of progressive myoclonus epilepsy:a multiprotein complex with cystatin B. Hum Mol Genet. 2002;11:2941-50. Links
-
67Lafrenière RG, Rochefort DL, Chrétien N, et al. Unstable insertion in the 5'flanking region of the cystatin B gene is the most common mutation in progressive myoclonus epilepsy Type 1, EPM1. Nat Genet. 1997;15:298-302. Links
-
68Lalioti MD, Mirotsou M, Buresi C, et al. Identification of mutations in cystatin B, the gene responsible for the unverricht-lundborg type of progressive myoclonus epilepsy (EPM1). Am J Hum Genet. 1997;60:342-51. Links
-
69Mazarib A, Xiong L, Neufeld MY, et al. Unverricht-lundborg disease in a five-generation arab family:instability of dodecamer repeats. Neurology. 2001;57:1050-4. Links
-
70Pennacchio LA, Lehesjoki AE, Stone NE, et al. Mutations in the gene encoding cystatin B in progressive myoclonus epilepsy (EPM1) Science. 1996;271:1731-4. Links
-
71Virtaneva K, D'Amato E, Miao J, et al. Unstable minisatellite expansion causing recessively inherited myoclonus epilepsy, EPM1. Nat Genet. 1997;15:393-6. Links
-
72Baulac S, Ishida S, Marsan E, et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann Neurol. 2015;77:675-83. Links
-
73Berkovic SF, Serratosa JM, Phillips HA, et al. Familial partial epilepsy with variable foci:clinical features and linkage to chromosome 22q12. Epilepsia. 2004;45:1054-60. Links
-
74Callenbach PM, van den Maagdenberg AM, Hottenga JJ, et al. Familial partial epilepsy with variable foci in a dutch family:clinical characteristics and confirmation of linkage to chromosome 22q. Epilepsia. 2003;44:1298-305. Links
-
75Dibbens LM, de Vries B, Donatello S, et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat Genet. 2013;45:546-51. Links
-
76Ishida S, Picard F, Rudolf G, et al. Mutations of DEPDC5 cause autosomal dominant focal epilepsies. Nat Genet. 2013;45:552-5. Links
-
77Klein KM, O'Brien TJ, Praveen K, et al. Familial focal epilepsy with variable foci mapped to chromosome 22q12:expansion of the phenotypic spectrum. Epilepsia. 2012;53:e151-5. Links
-
78Martin C, Meloche C, Rioux MF, et al. A recurrent mutation in DEPDC5 predisposes to focal epilepsies in the french-canadian population. Clin Genet. 2014;86:570-4. Links
-
79Picard F, Baulac S, Kahane P, et al. Dominant partial epilepsies. A clinical, electrophysiological and genetic study of 19 European families. Brain. 2000;123 (Pt 6):1247-62. Links
-
80Picard F, Makrythanasis P, Navarro V, et al. DEPDC5 mutations in families presenting as autosomal dominant nocturnal frontal lobe epilepsy. Neurology. 2014;82:2101-6. Links
-
81Scheffer IE, Heron SE, Regan BM, et al. Mutations in mammalian target of rapamycin regulator DEPDC5 cause focal epilepsy with brain malformations. Ann Neurol. 2014;75:782-7. Links
-
82Scheffer IE, Phillips HA, O'Brien CE, et al. Familial partial epilepsy with variable foci:a new partial epilepsy syndrome with suggestion of linkage to chromosome 2. Ann Neurol. 1998;44:890-9. Links
-
83Xiong L, Labuda M, Li DS, et al. Mapping of a gene determining familial partial epilepsy with variable foci to chromosome 22q11-q12. Am J Hum Genet. 1999;65:1698-710. Links
-
84Epilepsy Phenome/Genome Project Epi4K Consortium. Copy number variant analysis from exome data in 349 patients with epileptic encephalopathy. Ann Neurol. 2015;78:323-8. Links
-
85Boumil RM, Letts VA, Roberts MC, et al. A missense mutation in a highly conserved alternate exon of dynamin-1 causes epilepsy in fitful mice. PLoS Genet. 2010;6:e1001046. Links
-
86Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223-8. Links
-
87Dhindsa RS, Bradrick SS, Yao X, et al. Epileptic encephalopathy-causing mutations in DNM1 impair synaptic vesicle endocytosis. Neurol Genet. 2015;1:e4. Links
-
88EuroEPINOMICS-RES Consortium, Epilepsy Phenome/Genome Project, Epi4K Consortium. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet. 2014;95:360-70. Links
-
89Perrault I, Hamdan FF, Rio M, et al. Mutations in DOCK7 in individuals with epileptic encephalopathy and cortical blindness. Am J Hum Genet. 2014;94:891-7. Links
-
90Carvill GL, Weckhuysen S, McMahon JM, et al. GABRA1 and STXBP1:novel genetic causes of dravet syndrome. Neurology. 2014;82:1245-53. Links
-
91Cossette P, Liu L, Brisebois K, et al. Mutation of GABRA1 in an autosomal dominant form of juvenile myoclonic epilepsy. Nat Genet. 2002;31:184-9. Links
-
92Ding L, Feng HJ, Macdonald RL, et al. GABA(A) receptor alpha1 subunit mutation A322D associated with autosomal dominant juvenile myoclonic epilepsy reduces the expression and alters the composition of wild type GABA(A) receptors. J Biol Chem. 2010;285:26390-405. Links
-
93Lachance-Touchette P, Brown P, Meloche C, et al. Novel ?1 and ?2 GABAA receptor subunit mutations in families with idiopathic generalized epilepsy. Eur J Neurosci. 2011;34:237-49. Links
-
94Maljevic S, Krampfl K, Cobilanschi J, et al. A mutation in the GABA(A) receptor alpha(1)-subunit is associated with absence epilepsy. Ann Neurol. 2006;59:983-7. Links
-
95Tanaka M, Olsen RW, Medina MT, et al. Hyperglycosylation and reduced GABA currents of mutated GABRB3 polypeptide in remitting childhood absence epilepsy. Am J Hum Genet. 2008;82:1249-61. Links
-
96Urak L, Feucht M, Fathi N, Hornik K, Fuchs K. A GABRB3 promoter haplotype associated with childhood absence epilepsy impairs transcriptional activity. Hum Mol Genet. 2006;15:2533-41. Links
-
97Audenaert D, Schwartz E, Claeys KG, et al. A novel GABRG2 mutation associated with febrile seizures. Neurology. 2006;67:687-90. Links
-
98Baulac S, Huberfeld G, Gourfinkel-An I, et al. First genetic evidence of GABA(A) receptor dysfunction in epilepsy:a mutation in the gamma2-subunit gene. Nat Genet. 2001;28:46-8. Links
-
99Chaumont S, AndréC, Perrais D, Boué-Grabot E, et al. Agonist-dependent endocytosis of ?-aminobutyric acid Type A (GABAA) receptors revealed by a ?2(R43Q) epilepsy mutation. J Biol Chem. 2013;288:28254-65. Links
-
100Chiu C, Reid CA, Tan HO, et al. Developmental impact of a familial GABAA receptor epilepsy mutation. Ann Neurol. 2008;64:284-93. Links
-
101Frugier G, Coussen F, Giraud MF, et al. A gamma 2(R43Q) mutation, linked to epilepsy in humans, alters GABAA receptor assembly and modifies subunit composition on the cell surface. J Biol Chem. 2007;282:3819-28. Links
-
102Harkin LA, Bowser DN, Dibbens LM, et al. Truncation of the GABA(A)-receptor gamma2 subunit in a family with generalized epilepsy with febrile seizures plus. Am J Hum Genet. 2002;70:530-6. Links
-
103Kananura C, Haug K, Sander T, et al. A splice-site mutation in GABRG2 associated with childhood absence epilepsy and febrile convulsions. Arch Neurol. 2002;59:1137-41. Links
-
104Kang JQ, Shen W, Macdonald RL. Why does fever trigger febrile seizures?GABAA receptor gamma2 subunit mutations associated with idiopathic generalized epilepsies have temperature-dependent trafficking deficiencies. J Neurosci. 2006;26:2590-7. Links
-
105Sancar F, Czajkowski C. A GABAA receptor mutation linked to human epilepsy (gamma2R43Q) impairs cell surface expression of alphabetagamma receptors. J Biol Chem. 2004;279:47034-9. Links
-
106Tan HO, Reid CA, Single FN, et al. Reduced cortical inhibition in a mouse model of familial childhood absence epilepsy. Proc Natl Acad Sci U S A. 2007;104:17536-41. Links
-
107Wallace RH, Marini C, Petrou S, et al. Mutant GABA(A) receptor gamma2-subunit in childhood absence epilepsy and febrile seizures. Nat Genet. 2001;28:49-52. Links
-
108Nakamura K, Kodera H, Akita T, et al. De novo mutations in GNAO1, encoding a g?o subunit of heterotrimeric G proteins, cause epileptic encephalopathy. Am J Hum Genet. 2013;93:496-505. Links
-
109Carvill GL, Regan BM, Yendle SC, et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet. 2013;45:1073-6. Links
-
110Endele S, Rosenberger G, Geider K, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021-6. Links
-
111Lesca G, Rudolf G, Bruneau N, et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet. 2013;45:1061-6. Links
-
112Lemke JR, Lal D, Reinthaler EM, et al. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat Genet. 2013;45:1067-72. Links
-
113Scheffer IE, Jones L, Pozzebon M, et al. Autosomal dominant rolandic epilepsy and speech dyspraxia:a new syndrome with anticipation. Ann Neurol. 1995;38:633-42. Links
-
114Nava C, Dalle C, Rastetter A, et al. De novo mutations in HCN1 cause early infantile epileptic encephalopathy. Nat Genet. 2014;46:640-5. Links
-
115Bassi MT, Balottin U, Panzeri C, et al. Functional analysis of novel KCNQ2 and KCNQ3 gene variants found in a large pedigree with benign familial neonatal convulsions (BFNC). Neurogenetics. 2005;6:185-93. Links
-
116Berkovic SF, Kennerson ML, Howell RA, et al. Phenotypic expression of benign familial neonatal convulsions linked to chromosome 20. Arch Neurol. 1994;51:1125-8. Links
-
117Biervert C, Schroeder BC, Kubisch C, et al. A potassium channel mutation in neonatal human epilepsy. Science. 1998;279:403-6. Links
-
118Biervert C, Steinlein OK. Structural and mutational analysis of KCNQ2, the major gene locus for benign familial neonatal convulsions. Hum Genet. 1999;104:234-40. Links
-
119Borgatti R, Zucca C, Cavallini A, et al. A novel mutation in KCNQ2 associated with BFNC, drug resistant epilepsy, and mental retardation. Neurology. 2004;63:57-65. Links
-
120Dedek K, Fusco L, Teloy N, Steinlein OK. Neonatal convulsions and epileptic encephalopathy in an italian family with a missense mutation in the fifth transmembrane region of KCNQ2. Epilepsy Res. 2003;54:21-7. Links
-
121del Giudice EM, Coppola G, Scuccimarra G, et al. Benign familial neonatal convulsions (BFNC) resulting from mutation of the KCNQ2 voltage sensor. Eur J Hum Genet. 2000;8:994-7. Links
-
122Heron SE, Cox K, Grinton BE, et al. Deletions or duplications in KCNQ2 can cause benign familial neonatal seizures. J Med Genet. 2007;44:791-6. Links
-
123Saitsu H, Kato M, Koide A, et al. Whole exome sequencing identifies KCNQ2 mutations in ohtahara syndrome. Ann Neurol. 2012;72:298-300. Links
-
124Singh NA, Charlier C, Stauffer D, et al. A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nat Genet. 1998;18:25-9. Links
-
125Weckhuysen S, Mandelstam S, Suls A, et al. KCNQ2 encephalopathy:emerging phenotype of a neonatal epileptic encephalopathy. Ann Neurol. 2012;71:15-25. Links
-
126Wuttke TV, Jurkat-Rott K, Paulus W, et al. Peripheral nerve hyperexcitability due to dominant-negative KCNQ2 mutations. Neurology. 2007;69:2045-53. Links
-
127Yang WP, Levesque PC, Little WA, et al. Functional expression of two kvLQT1-related potassium channels responsible for an inherited idiopathic epilepsy. J Biol Chem. 1998;273:19419-23. Links
-
128Zimprich F, Ronen GM, Stögmann W, et al. Andreas rett and benign familial neonatal convulsions revisited. Neurology. 2006;67:864-6. Links
-
129Barcia G, Fleming MR, Deligniere A, et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat Genet. 2012;44:1255-9. Links
-
130Derry CP, Heron SE, Phillips F, et al. Severe autosomal dominant nocturnal frontal lobe epilepsy associated with psychiatric disorders and intellectual disability. Epilepsia. 2008;49:2125-9. Links
-
131Heron SE, Smith KR, Bahlo M, et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 2012;44:1188-90. Links
-
132Ishii A, Shioda M, Okumura A, et al. A recurrent KCNT1 mutation in two sporadic cases with malignant migrating partial seizures in infancy. Gene. 2013;531:467-71. Links
-
133Ohba C, Kato M, Takahashi S, et al. Early onset epileptic encephalopathy caused by de novo SCN8A mutations. Epilepsia. 2014;55:994-1000. Links
-
134Vanderver A, Simons C, Schmidt JL, et al. Identification of a novel de novo p.Phe932Ile KCNT1 mutation in a patient with leukoencephalopathy and severe epilepsy. Pediatr Neurol. 2014;50:112-4. Links
-
135Chabrol E, Popescu C, Gourfinkel-An I, et al. Two novel epilepsy-linked mutations leading to a loss of function of LGI1. Arch Neurol. 2007;64:217-22. Links
-
136Fanciulli M, Santulli L, Errichiello L, et al. LGI1 microdeletion in autosomal dominant lateral temporal epilepsy. Neurology. 2012;78:1299-303. Links
-
137Fertig E, Lincoln A, Martinuzzi A, Mattson RH, Hisama FM. Novel LGI1 mutation in a family with autosomal dominant partial epilepsy with auditory features. Neurology. 2003;60:1687-90. Links
-
138Kalachikov S, Evgrafov O, Ross B, et al. Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat Genet. 2002;30:335-41. Links
-
139Morante-Redolat JM, Gorostidi-Pagola A, Piquer-Sirerol S, et al. Mutations in the LGI1/Epitempin gene on 10q24 cause autosomal dominant lateral temporal epilepsy. Hum Mol Genet. 2002;11:1119-28. Links
-
140Nobile C, Michelucci R, Andreazza S, et al. LGI1 mutations in autosomal dominant and sporadic lateral temporal epilepsy. Hum Mutat. 2009;30:530-6. Links
-
141Sirerol-Piquer MS, Ayerdi-Izquierdo A, Morante-Redolat JM, et al. The epilepsy gene LGI1 encodes a secreted glycoprotein that binds to the cell surface. Hum Mol Genet. 2006;15:3436-45. Links
-
142Striano P, de Falco A, Diani E, et al. A novel loss-of-function LGI1 mutation linked to autosomal dominant lateral temporal epilepsy. Arch Neurol. 2008;65:939-42. Links
-
143Depienne C, Bouteiller D, Keren B, et al. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles dravet syndrome but mainly affects females. PLoS Genet. 2009;5:e1000381. Links
-
144Dibbens LM, Tarpey PS, Hynes K, et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008;40:776-81. Links
-
145Hynes K, Tarpey P, Dibbens LM, et al. Epilepsy and mental retardation limited to females with PCDH19 mutations can present de novo or in single generation families. J Med Genet. 2010;47:211-6. Links
-
146Kurian MA, Meyer E, Vassallo G, et al. Phospholipase C beta 1 deficiency is associated with early-onset epileptic encephalopathy. Brain. 2010;133:2964-70. Links
-
147Poduri A, Chopra SS, Neilan EG, et al. Homozygous PLCB1 deletion associated with malignant migrating partial seizures in infancy. Epilepsia. 2012;53:e146-50. Links
-
148Shen J, Gilmore EC, Marshall CA, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245-9. Links
-
149Chen WJ, Lin Y, Xiong ZQ, et al. Exome sequencing identifies truncating mutations in PRRT2 that cause paroxysmal kinesigenic dyskinesia. Nat Genet. 2011;43:1252-5. Links
-
150Heron SE, Grinton BE, Kivity S, et al. PRRT2 mutations cause benign familial infantile epilepsy and infantile convulsions with choreoathetosis syndrome. Am J Hum Genet. 2012;90:152-60. Links
-
151Lee HY, Huang Y, Bruneau N, et al. Mutations in the gene PRRT2 cause paroxysmal kinesigenic dyskinesia with infantile convulsions. Cell Rep. 2012;1:2-12. Links
-
152Méneret A, Grabli D, Depienne C, et al. PRRT2 mutations:a major cause of paroxysmal kinesigenic dyskinesia in the European population. Neurology. 2012;79:170-4. Links
-
153Ono S, Yoshiura K, Kinoshita A, et al. Mutations in PRRT2 responsible for paroxysmal kinesigenic dyskinesias also cause benign familial infantile convulsions. J Hum Genet. 2012;57:338-41. Links
-
154Pelzer N, de Vries B, Kamphorst JT, et al. PRRT2 and hemiplegic migraine:a complex association. Neurology. 2014;83:288-90. Links
-
155Schubert J, Paravidino R, Becker F, et al. PRRT2 mutations are the major cause of benign familial infantile seizures. Hum Mutat. 2012;33:1439-43. Links
-
156Striano P, Lispi ML, Gennaro E, et al. Linkage analysis and disease models in benign familial infantile seizures:a study of 16 families. Epilepsia. 2006;47:1029-34. Links
-
157Wang JL, Cao L, Li XH, Hu ZM, Li JD, Zhang JG, et al. Identification of PRRT2 as the causative gene of paroxysmal kinesigenic dyskinesias. Brain. 2011;134:3493-501. Links
-
158Weber YG, Berger A, Bebek N, et al. Benign familial infantile convulsions:linkage to chromosome 16p12-q12 in 14 families. Epilepsia. 2004;45:601-9. Links
-
159Abou-Khalil B, Ge Q, Desai R, et al. Partial and generalized epilepsy with febrile seizures plus and a novel SCN1A mutation. Neurology. 2001;57:2265-72. Links
-
160Baulac S, Gourfinkel-An I, Picard F, et al. A second locus for familial generalized epilepsy with febrile seizures plus maps to chromosome 2q21-q33. Am J Hum Genet. 1999;65:1078-85. Links
-
161Buoni S, Orrico A, Galli L, et al. SCN1A (2528delG) novel truncating mutation with benign outcome of severe myoclonic epilepsy of infancy. Neurology. 2006;66:606-7. Links
-
162Rojo DC, Hamiwka L, McMahon JM, et al. De novo SCN1A mutations in migrating partial seizures of infancy. Neurology. 2011;77:380-3. Links
-
163Claes L, Del-Favero J, Ceulemans B, et al. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68:1327-32. Links
-
164Dichgans M, Freilinger T, Eckstein G, et al. Mutation in the neuronal voltage-gated sodium channel SCN1A in familial hemiplegic migraine. Lancet. 2005;366:371-7. Links
-
165Escayg A, MacDonald BT, Meisler MH, et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343-5. Links
-
166Freilich ER, Jones JM, Gaillard WD, et al. Novel SCN1A mutation in a proband with malignant migrating partial seizures of infancy. Arch Neurol. 2011;68:665-71. Links
-
167Mantegazza M, Gambardella A, Rusconi R, et al. Identification of an nav1.1 sodium channel (SCN1A) loss-of-function mutation associated with familial simple febrile seizures. Proc Natl Acad Sci U S A. 2005;102:18177-82. Links
-
168McArdle EJ, Kunic JD, George AL Jr. Novel SCN1A frameshift mutation with absence of truncated nav1.1 protein in severe myoclonic epilepsy of infancy. Am J Med Genet A. 2008;146A:2421-3. Links
-
169Moulard B, Buresi C, Malafosse A. Study of the voltage-gated sodium channel beta 1 subunit gene (SCN1B) in the benign familial infantile convulsions syndrome (BFIC). Hum Mutat. 2000;16:139-42. Links
-
170Mulley JC, Scheffer IE, Petrou S, et al. SCN1A mutations and epilepsy. Hum Mutat. 2005;25:535-42. Links
-
171Ohmori I, Ouchida M, Ohtsuka Y, Oka E, Shimizu K. Significant correlation of the SCN1A mutations and severe myoclonic epilepsy in infancy. Biochem Biophys Res Commun. 2002;295:17-23. Links
-
172Orrico A, Galli L, Grosso S, et al. Mutational analysis of the SCN1A, SCN1B and GABRG2 genes in 150 italian patients with idiopathic childhood epilepsies. Clin Genet. 2009;75:579-81. Links
-
173Petrovski S, Scheffer IE, Sisodiya SM, et al. Lack of replication of association between scn1a SNP and febrile seizures. Neurology. 2009;73:1928-30. Links
-
174Schlachter K, Gruber-Sedlmayr U, Stogmann E, et al. A splice site variant in the sodium channel gene SCN1A confers risk of febrile seizures. Neurology. 2009;72:974-8. Links
-
175Vahedi K, Depienne C, Le Fort D, et al. Elicited repetitive daily blindness:a new phenotype associated with hemiplegic migraine and SCN1A mutations. Neurology. 2009;72:1178-83. Links
-
176Zucca C, Redaelli F, Epifanio R, et al. Cryptogenic epileptic syndromes related to SCN1A:twelve novel mutations identified. Arch Neurol. 2008;65:489-94. Links
-
177Berkovic SF, Heron SE, Giordano L, et al. Benign familial neonatal-infantile seizures:characterization of a new sodium channelopathy. Ann Neurol. 2004;55:550-7. Links
-
178Heron SE, Crossland KM, Andermann E, et al. Sodium-channel defects in benign familial neonatal-infantile seizures. Lancet. 2002;360:851-2. Links
-
179Kamiya K, Kaneda M, Sugawara T, et al. A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. J Neurosci. 2004;24:2690-8. Links
-
180Liao Y, Anttonen AK, Liukkonen E, et al. SCN2A mutation associated with neonatal epilepsy, late-onset episodic ataxia, myoclonus, and pain. Neurology. 2010;75:1454-8. Links
-
181Malacarne M, Gennaro E, Madia F, et al. Benign familial infantile convulsions:mapping of a novel locus on chromosome 2q24 and evidence for genetic heterogeneity. Am J Hum Genet. 2001;68:1521-6. Links
-
182Ogiwara I, Ito K, Sawaishi Y, et al. De novo mutations of voltage-gated sodium channel alphaII gene SCN2A in intractable epilepsies. Neurology. 2009;73:1046-53. Links
-
183Sugawara T, Tsurubuchi Y, Agarwala KL, et al. A missense mutation of the na+channel alpha II subunit gene na(v)1.2 in a patient with febrile and afebrile seizures causes channel dysfunction. Proc Natl Acad Sci U S A. 2001;98:6384-9. Links
-
184Blanchard MG, Willemsen MH, Walker JB, et al. De novo gain-of-function and loss-of-function mutations of SCN8A in patients with intellectual disabilities and epilepsy. J Med Genet. 2015;52:330-7. Links
-
185de Kovel CG, Meisler MH, Brilstra EH, et al. Characterization of a de novo SCN8A mutation in a patient with epileptic encephalopathy. Epilepsy Res. 2014;108:1511-8. Links
-
186Veeramah KR, O'Brien JE, Meisler MH, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012;90:502-10. Links
-
187Arsov T, Mullen SA, Rogers S, et al. Glucose transporter 1 deficiency in the idiopathic generalized epilepsies. Ann Neurol. 2012;72:807-15. Links
-
188Striano P, Weber YG, Toliat MR, et al. GLUT1 muations are a rare cause of familial idiopathic generalized epilepsy. Neurology. 2012;78:557-62. Links
-
189Suls A, Mullen SA, Weber YG, et al. Early-onset absence epilepsy caused by mutations in the glucose transporter GLUT1. Ann Neurol. 2009;66:415-9. Links
-
190Molinari F, Raas-Rothschild A, Rio M, et al. Impaired mitochondrial glutamate transport in autosomal recessive neonatal myoclonic epilepsy. Am J Hum Genet. 2005;76:334-9. Links
-
191Poduri A, Heinzen EL, Chitsazzadeh V, et al. SLC25A22 is a novel gene for migrating partial seizures in infancy. Ann Neurol. 2013;74:873-82. Links
-
192Hamdan FF, Saitsu H, Nishiyama K, et al. Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy. Eur J Hum Genet. 2012;20:796-800. Links
-
193Nonoda Y, Saito Y, Nagai S, et al. Progressive diffuse brain atrophy in west syndrome with marked hypomyelination due to SPTAN1 gene mutation. Brain Dev. 2013;35:280-3. Links
-
194Saitsu H, Tohyama J, Kumada T, et al. Dominant-negative mutations in alpha-II spectrin cause west syndrome with severe cerebral hypomyelination, spastic quadriplegia, and developmental delay. Am J Hum Genet. 2010;86:881-91. Links
-
195Hamdan FF, Piton A, Gauthier J, et al. De novo STXBP1 mutations in mental retardation and nonsyndromic epilepsy. Ann Neurol. 2009;65:748-53. Links
-
196Saitsu H, Kato M, Mizuguchi T, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet. 2008;40:782-8. Links
-
197Edvardson S, Baumann AM, Mühlenhoff M, et al. West syndrome caused by ST3Gal-III deficiency. Epilepsia. 2013;54:e24-7. Links
-
198Berryer MH, Hamdan FF, Klitten LL, et al. Mutations in SYNGAP1 cause intellectual disability, autism, and a specific form of epilepsy by inducing haploinsufficiency. Hum Mutat. 2013;34:385-94. Links
-
199Mignot C, von Stülpnagel C, Nava C, et al. Genetic and neurodevelopmental spectrum of SYNGAP1-associated intellectual disability and epilepsy. J Med Genet. 2016;53:511-22. Links
-
200Basel-Vanagaite L, Hershkovitz T, Heyman E, et al. Biallelic SZT2 mutations cause infantile encephalopathy with epilepsy and dysmorphic corpus callosum. Am J Hum Genet. 2013;93:524-9. Links
-
201Corbett MA, Bahlo M, Jolly L, et al. A focal epilepsy and intellectual disability syndrome is due to a mutation in TBC1D24. Am J Hum Genet. 2010;87:371-5. Links
-
202Duru N, Iseri SA, Selçuk N, Tolun A. Early-onset progressive myoclonic epilepsy with dystonia mapping to 16pter-p13.3. J Neurogenet. 2010;24:207-15. Links
-
203Falace A, Filipello F, La Padula V, et al. TBC1D24, an ARF6-interacting protein, is mutated in familial infantile myoclonic epilepsy. Am J Hum Genet. 2010;87:365-70. Links
-
204Guven A, Tolun A. TBC1D24 truncating mutation resulting in severe neurodegeneration. J Med Genet. 2013;50:199-202. Links
-
205Milh M, Falace A, Villeneuve N, et al. Novel compound heterozygous mutations in TBC1D24 cause familial malignant migrating partial seizures of infancy. Hum Mutat. 2013;34:869-72. Links
-
206Zara F, Gennaro E, Stabile M, et al. Mapping of a locus for a familial autosomal recessive idiopathic myoclonic epilepsy of infancy to chromosome 16p13. Am J Hum Genet. 2000;66:1552-7. Links
Supplementary material
Supplementary Table 1
Epilepsy related genes
Epilepsy related genes
| Gen | Proteina | Localización | Tipo | Alteración | Fenotipo | Técnica | Fuentes |
|---|---|---|---|---|---|---|---|
| ALG13 | Asparagine-linked glycosylation 13, S. cerevisiae, homolog of glycosyltransferase 28 domain-containing 1 | Xq23 | SNP | - LYS94GLU |
EETI, EI | WES | De Ligt et al., 20121 |
| ARHGEF9 | Rho guanine nucleotide exchange factor 9 (Collybistin) | Xq11 | SNP | - GLY55ALA |
EIEE | Array CGH | Harvey et al., 20047 |
| ARX | Aristaless-related homeobox, X-Linked | Xp21.3 | Dup |
- 24-BP DUP, NT428 |
EEIT | CGH |
Bruyere et al., 199911
|
| CACNA1A | Subunidad alpha-1-a de canal P/Q de calcio dependiente de voltaje | 19p13.13 | SNP | - ARG1820TER | Aus, EGI | WES |
Chan et al., 200823
|
| CACNA1H | Calcium channel, voltage-dependent, T type, alpha 1H subunit | 16p13.3 | SNP | - PHE161LEU |
Aus, EGI | DES |
Chen et al., 200328
|
| CACNA2D1 | Calcium channel, voltage-dependent. alpha2/delta subunit 1 | 7q11-q21 | Del | - 7.5-MB deletion 7q21.11-q21.12 |
ESG | GWEF Array CGH | Mefford et al., 201133
|
| CDKL5 | Cyclin-dependent kinase-like 5 | Xp22.13 | Del SNP | - 1-BP DEL, 183T |
EEIT, EI, EMT | WES |
Archer et al., 200635
|
| CHD2 | Chromodomain helicase DNA-binding protein 2 | 15q26.1 | Del |
- 1-BP DEL, 1809G GLU1412GLYFSTER64 |
Síndrome de Dravet, Lennox-Gastaut y Doose. (Fotosensible) EMA, Aus | WES, Targeted sequencing | Carvill et al., 2013,50
|
| CHRNA7 | Cholinergic receptor, nicotinic, alpha 7 | 15q13.3 | Del | EGG | GWAS |
Dibbens et al., 200959
|
|
| CNTNAP2 | Contactin-associated protein-like 2 | 7q35 | Del | 1-BP DEL, 3709G | EF con regresión. Síndrome Epilepsia Focal-Displasia Cortical | GWEF array CGH Targeted sequencing | Mefford et al., 201133
|
| CSTB | Cystatin-B | 21q22.3 | Del SNP | - IVS1, G-C, -1 |
Epilepsia mioclónica progresiva (Síndrome de Unverricht-Lundborg) | Complete sequencing of the gene | Alakurtti et al., 200563
|
| DEPDC5 | Dominio DEP 5 | 22q12.3 | Del |
- TYR7TER |
EFFFM, ELFNAD, ELTMF | WES, Direct sequencing | Baulac et al., 201572
|
| DMRT2, DMRT3 | Doublesex- and Mab-3-related transcription factor 2 and factor 3 | 9p24.3 | Del | EI | Array - CGH | Epi4K Consortium and Epilepsy Phenome/Genome Project, 201584 | |
| DNM1 | Dynamin 1 | 9q34.11 | SNP | ALA177PRO |
EEIT (Lennox-Gastaut), EI | WES | Møller, 20155
|
| DOCK7 | Dedicator for cytokinesis 7 | 1p31.3 | Del |
- 1-BP DEL, 2510A |
EEIT | WES | Perrault et al., 201489 |
| GABRA1 | Gamma-aminobutyric acid (GABA) A receptor, alpha 1 | 5q34 | SNP |
- ALA322ASP |
EEIT, EMJ, Aus | Array - CGH |
Carvill et al., 201490
|
| GABRB3 | Gamma-aminobutyric acid A receptor, beta 3 | 15q11 | SNP | - PRO11SER |
EI, TCG, T, atonicas, Aus | Array - CGH |
Epi4K Consortium and Epilepsy Phenome/Genome
Project, 201584
|
| GABRG2 | Receptor GABA-A, Polipéptido gamma-2 | 5q34 | SNP | - LYS289MET |
EGI, CF, Aus | Candidate gene sequencing | Audenaert et al., 200697
|
| GNAO1 | Guanine nucleotide-binding protein alpha activating | 16q12.2 | SNP |
- ILE279ASN |
EEIT | CGH |
Lesca, 20159
|
| GRIN2A | Glutamate receptor, ionotropic, N-methyl D-aspartate 2A | 16p13.2 | SNP | GLN218TER IVS4DS, G-A, +1 ASN615LYS LEU649VAL PRO522ARG MET1THR THR531MET IVS5AS, A-G, -2 ARG518HIS PHE652VAL ARG681TER TYR943TER | SEA, EF | WES | Møller, 20155
|
| HCN1 | Hyperpolarization-activated cyclic nucleotide-gated potassium channel 1 | 5p12 | SNP | ASP401HIS |
EEIT | CGH |
Lesca, 20159
|
| HDAC4 | Histone deacetylase 4 | 2q37.3 | EEIT | WES | Møller, 20155 | ||
| HIP1 | Huntingtin interacting Protein 1 | 7q11 | EI | Array - CGH | Epi4K Consortium and Epilepsy Phenome/Genome Project, 201584 | ||
| KCNQ2 | Potassium channel, voltage-gated, KQT-like subfamily, member 2 | 20q13.3 | SNP |
TYR284CYS |
EEIT, ENFB | CGH |
Lesca, 20159
|
| KCNT1 | Potassium channel, sodium-activated subfamily T, member 1 | 9q34.4 | SNP | ARG428GLN |
ELFNAD, EICFM | WES | Barcia et al., 2012129
|
| LGI1 | Leucine-rich gene, glioma inactivated 1 | 10q23.33 | SNP |
GLU383ALA |
ELTMF | Direct sequencing CNV analysis | Chabrol et al., 2007135
|
| PCDH19 | Protocadherin 19 | Xq22.1 | SNP |
1-BP INS, 1091C |
EEIT en mujeres (Ohtahara, Dravet) | Microarrays, Systematic resequencing | Depienne et al., 2009143
|
| PLCB1 | Phospholipase C, beta-1 | 20p12.3 | Del | 0.5-MB DEL | EEIT, CPMMI | CGH Genome-wide scan | Lesca, 20159
|
| PNKP | Polynucleotide kinase 3 phosphatase | 19q13.33 | SNP |
GLU326LYS |
EEIT, Microcefalia-Crisis y Retraso Mental | CGH Genome-wide scan | Lesca, 20159
|
| PRRT2 | Proline-rich transmembrane protein 2 | 16p11.2 | Dup |
1-BP DUP, 649C |
CIF, CICA | WES | Møller, 20155
|
| RYR3 | Ryanodine receptor 3 | 15q13.3 | EEIT | WES | Møller, 20155 | ||
| SCN1A | Sodium voltage-gated channel alpha subunit 1 | 2q24.3 | SNP |
ARG1648HIS |
Síndrome de Dravet, CF familiares, EEIT | WES |
Abou-Khalil et al., 2001159
|
| SCN2A | Sodium channel, voltage-gated, type II, alpha subunit | 2q24.3 | ARG188TRP |
EEIT | Direct sequencing, WES, Genome-wide analysis. | Berkovic et al., 2004177
|
|
| SCN8A | Sodium Channel, voltage-gated, type VIII, alpha subunit | 12q13.13 | SNP | ASN1768ASP |
EEIT |
WES |
Blanchard et al., 2015184
|
| SLC2A1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | 1p34.2 | SNP | ARG232CYS |
EGI | Direct sequencing, PCR sequencing | Arsov et al., 2012187
|
| SLC25A22 | Solute carrier family 25 (mitochondrial carrier, glutamate) member 22 | 11p15.5 | SNP | PRO206LEU |
EEIT | CGH |
Lesca, 20159
|
| SLC26A1 | Solute carrier family 26, (anion exchanger), member 1 | 4p16 | SEA | GWEF array CGH | Mefford et al., 201130 | ||
| SLC35A2 | Solute carrier family 35 (UDP-galactose transporter) member 2 | Xp11.23 | Del SNP | 2-BP DEL, 433TA |
EEIT | CGH |
Lesca, 20159
|
| SPTAN1 | Alpha, non-erythrocytic, spectrin 1 | 9q34.11 | Del Dup | 3-BP DEL, 6619GAG |
EEIT | CGH |
Lesca, 20159
|
| STXBP1 | Syntaxin-binding protein 1 | 9q34.11 | SNP | GLY544ASP |
EIEE | WES | Møller, 20155
|
| STX1B | Syntaxin 1B | 16p11.2 | Síndromes asociados con epilepsia febril | WES | Møller, 20155 | ||
| ST3GAL3 | ST3 beta-galactoside alpha-2,3-sialyltransferase 3 | 1p34.1 | SNP | ALA320PRO | EEIT | CGH |
Lesca, 20159
|
| SYNGAP1 | Synaptic RAS-GTPase-activating protein 1 | 6p21.32 | SNP | PRO562LEU |
EEIT, mioclónicas, Aus | WES | Barryer et al., 2013198
|
| SZT2 | Seizure threshold 2, mouse homolog | 1p34.2 | SNP | ARG25TER |
EEIT | CGH |
Basel-Vanagaite et al., 2013200
|
| TAS2R1, FAM173B, CCT5, MTRR | Taste receptor, type 2, member 1/family with sequence similarity 173, member B/chemokine receptor 5/5- methyltetrahydrofolate-homocysteine methyltransferase reductase | 5p15 | FS, focal, TCG, aA, SE | Array - CGH | Epi4K Consortium and Epilepsy Phenome/Genome Project, 201584 | ||
| TBC1D24 | TBC1 domain family, member 24 | 16p13.3 | SNP |
ASP147HIS |
EEIT, EMIF | CGH |
Lesca, 20159
|
| UBE3A | Ubiquitin protein ligase E3A | 15q11 | Del | EMA | GWEF array CGH | Mefford et al., 201133 |